+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10232 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the closed conformation of CtTel1 | |||||||||
 Map data Map data | One protomer in closed conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / alpha solenoid / PIKK / nucleus / tranferase / dimer / DNA damage signaling / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationchromatin organization / chromosome, telomeric region / non-specific serine/threonine protein kinase / intracellular signal transduction / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) / Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) /  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Jansma M / Eustermann SE | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State. Authors: Marijke Jansma / Christian Linke-Winnebeck / Sebastian Eustermann / Katja Lammens / Dirk Kostrewa / Kristina Stakyte / Claudia Litz / Brigitte Kessler / Karl-Peter Hopfner /  Abstract: Tel1 (ATM in humans) is a large kinase that resides in the cell in an autoinhibited dimeric state and upon activation orchestrates the cellular response to DNA damage. We report the structure of an ...Tel1 (ATM in humans) is a large kinase that resides in the cell in an autoinhibited dimeric state and upon activation orchestrates the cellular response to DNA damage. We report the structure of an endogenous Tel1 dimer from Chaetomium thermophilum. Major parts are at 2.8 Å resolution, including the kinase active site with ATPγS bound, and two different N-terminal solenoid conformations are at 3.4 Å and 3.6 Å, providing a side-chain model for 90% of the Tel1 polypeptide. We show that the N-terminal solenoid has DNA binding activity, but that its movements are not coupled to kinase activation. Although ATPγS and catalytic residues are poised for catalysis, the kinase resides in an autoinhibited state. The PIKK regulatory domain acts as a pseudo-substrate, blocking direct access to the site of catalysis. The structure allows mapping of human cancer mutations and defines mechanisms of autoinhibition at near-atomic resolution. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10232.map.gz emd_10232.map.gz | 12.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10232-v30.xml emd-10232-v30.xml emd-10232.xml emd-10232.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10232_fsc.xml emd_10232_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_10232.png emd_10232.png | 16.6 KB | ||
| Filedesc metadata |  emd-10232.cif.gz emd-10232.cif.gz | 7.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10232 http://ftp.pdbj.org/pub/emdb/structures/EMD-10232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10232 | HTTPS FTP |
-Related structure data
| Related structure data |  6skzMC  6skyC  6sl0C  6sl1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10232.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10232.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One protomer in closed conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
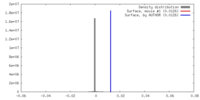
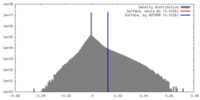
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CtTel1
| Entire | Name: CtTel1 |
|---|---|
| Components |
|
-Supramolecule #1: CtTel1
| Supramolecule | Name: CtTel1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 640 KDa |
-Macromolecule #1: Serine/threonine-protein kinase Tel1
| Macromolecule | Name: Serine/threonine-protein kinase Tel1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 329.927031 KDa |
| Sequence | String: MPRVKSYTGG TINFKEIESH LVGSSATARK DAVEALIAYF DGSRTQSQTT FDDKAYHRLF EALFRCTLIE KEAYFNSKKS VKVTAAAAA RLERCPEALR LAVRHGVTTI RRKTARAIID HIVQVLPGPD GAYVMPLLAG YVKVLYEFLD NPASAENIAA L SGEGWEVC ...String: MPRVKSYTGG TINFKEIESH LVGSSATARK DAVEALIAYF DGSRTQSQTT FDDKAYHRLF EALFRCTLIE KEAYFNSKKS VKVTAAAAA RLERCPEALR LAVRHGVTTI RRKTARAIID HIVQVLPGPD GAYVMPLLAG YVKVLYEFLD NPASAENIAA L SGEGWEVC VDFCIDVLSR FLELGDRESG SLSRASPAPG ATARSGSVAG TQGSGEQIGT HVAVDVLSCL YMLCIAHNAP IQ RKADRLP HVVIQLLQLR QMKIGELQKM AFATFNIVFQ RMQAEDVALC KTLVKQVVPL LSHWWQPRAL SRDAMLNSIR DEM LKTLYG TRLYIQALLR EAADESFPQD VEELLDTLWC DYSRREERAR LQLDDITFTN MLLPPDHPRT GIFSLRPHHT AGEQ NWALL ENLAILEAAY SKHGQQEQSQ QNQQQPEIDQ PRKRRKMSGR QNRVHQKLHS LDPAVRLSAL QLIPFLTRHK KPSLE DVAE TLEDLSKHVT AKQAIVASWA MLACSSLAIH EVSRHPSLSS SWKQLWQLAV RSLSLPPISR ASCVLLNSIL KANLIP RHE LADDINQIVT TADISGPAIL VDASLGLMLN LLRFRNNMFP NASQATSNHI IRWVFVRWSP AELTYASLHG THATPYD LV NLLRACYGIS PLVMAQPLRL FNGPIVLHWK EQAEMEPFIR YLLLLHEEEP DTTVTPAQQE EQLPESNSAT DVAGSNAS R RLALELFYPK VEELQELAES WQKRGGEGAT PVSMERLRSM VLACLTGALL LPDLVNINSS LSRDLESAVF SIVDATLKV ILNSPPSENL FGMILASSAP YIPHLIEPEL IALKRERPHL LKFFGKLSEA LYERSRRESS HRDDQVIDID PDFEPQTSQK NTASKAKTL PRRDILLSYT PEAFYLETSL RIHFLDIIRL NDGEIGRIPE PIINQLAGLS GEQFLCCREF MREIFTSDAI V PLGGATTI LETAGHIVSR YEYACCEVAL CNCIDIMDSF INLWTDNHFD IAEMAGDLYH YLVKQSLPNN SMSAAAQIRL AS LLLHLLE VKSEYASNLG LPSSQSTLLK ILQDGPMKLK HYIGLEIPKL FGLYVLKTHD DIFVDVLEHL PSDPDVVEGL AFR LFVLAE LACRWPTLLR RSIYHTFEIP GKITKISKSQ SCVTHSALYA ASCIKRIAQT LKLSGPQELF KLFAPQLLYT WLDN DSIQD IAYEIFGFSS LLDLLREAQT EAAAIMMMRG QEQEVCQLAQ SLGLTPEKLV QQSFTKIIAY SIAHDISIAG GPDYV TGES RMRKILGKEE YLANIHLNFA DIISTFFDIF DQEDPIEKAF RRDERFAYAA ETLEEIKKLG HLPTALPPNQ QPMFRA KYL PREIVHLCSR TQYEPENIWT PALVVFVARK LLKTIHPALG PLHACSVLRK IRVLICLAGD HAISGYPLEM LLHSLRV FV VDPECADDAL GITQYLIKRG DEYLKRTPSF LAGYALSSLA DLRVFLESSQ SSTTQESQFK ATKSKAQEFH AWFSKYLA A YDSPEFKDEG QKQAFRSITE NAAHIRASGN AEKGTHESNL LLEILKDWGR ENQLLNEPAR DVALSMLCGV FNIPPSSRL DVIETDEDAI KNGAVVWKSC SSQRLGGEYL AWAGRVLGRS FAASGEVPED LLRESQLQEY RRLSQGVGSS EEGLLNLIKS LTISGDCFT AGLAEAALRT IVSDAISDND HDLLSACQES LPEPLLIASN WDPYRTPLSD QFKVDPPANT EVFSARALEN P NWSQHLAI RLALSAPKIV TLRVLPPILS KVKGFAERAF PFVVHLVLAY QLDKQQSAKR ELSESLQEWL NFTSEPAKEN LK LLINTIL YLRTQPLPGE SSIADRAHWL DVNMASAAAA ATRCGMYKVA LLFAELAAES TRGSRRSSAA RETDDSSDIL LEI FENIDD PDAYYGLSQD ASLSTVLARL EYENDGAKSL AFRGAQYDSH LRGRDLQSRQ DCNALIKALS SLGLAGLSNS LLQS QQSID GSSDSLDATF TTARKLEIWN LPAPVNSDSW AVTVYKAYQS MYQAQELDTV RSMVHDGLKN TVRHLSSGSL NTSVL RQQL GALAALTELD DILNVRDQSE LQCTLATFEK RSKWMMSGRY ADVSQILSCR ETTLSMWSQR HNLRAAGLTS ADARLV QIR GMLLSSDIFR FHRARQETLN LSTALSDLIP SCESLGLSVD AAIKMEAANA LWDHGEMISS IRMLQAIDKD SSLKKQS VP LSRSDLLSKI GYQVSVARLE SPDAIQKKYL EPALKELKGK IEGREAGQVF HQFAVFCDEQ LQNPDSLEDL ARLQNLKK G KDEEVAQLKA LIASAKDSQL RNRYQSHLAK AKQWQELDQQ ELRRVEQTRS EFLKLCIENY LLSLAASDEH DNDALRFMA LWLEKSEEEV ANEVVKKWIN KVPTRKFALL MNQLSSRLQD HNTLFQKLLI DLVYRICVDH PYHGMYHIWT GARTRVNKDD EVAVSRQRA TDKIAKALSK NNKVSSIWPA IDQTSRVYHA LAMDRDPTRY KSGQKVPIKN SPVGQNFLST MSNNPIPPPT L QIEVSANL DYSHVPMIHK FAPEMAIASG VSAPKILTAI GTDGRKYKQL VKGGNDDLRQ DAIMEQVFAA VSELLKLHRE TR QRNLGIR TYKVLPLTSS SGLIEFVSNT IPLHEYLMPA HERYYPKDLK GSQCRKEIAN AQTKNTETRI AVYRRVTERF HPV MRYFFM EYFPDPDEWF QKRTNYTRTT AAISMLGHVL GLGDRHGHNI LLDHKTGEVV HIDLGVAFEM GRVLPVPELV PFRL TRDIV DGMGITKTEG VFRRCCEFTL DALREEAASI QTILDSLRHD TLYQWSISPV RMAKLQNARE VGGEDGGVGG GEDGE GGGV PKEKKQRPAN EPSEADRAIE VVKKKLSKTL SVMATVNDLI NQATSVSNLA VLYSGWAAYA UniProtKB: Serine/threonine-protein kinase Tel1 |
-Macromolecule #2: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 1 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 1 mM MgCl2 and 0.1 mM ATPgS (final concentrations) added before plunging | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288.15 K / Instrument: LEICA EM GP Details: TWEEN-20 was added to a final concentration of 0.05% immediately before vitrification. Sample was preincubated 45 seconds before plunging into liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 13786 / Average exposure time: 8.0 sec. / Average electron dose: 55.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)