+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10210 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of bacterial flagellar capping protein FliD | |||||||||
 Map data Map data | FliDcj full model | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flagellum / Flagella / FliD / HAP2 / Campylobacter jejuni / C.jejuni / transport protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum filament cap / bacterial-type flagellum hook / cell adhesion / extracellular region Similarity search - Function | |||||||||
| Biological species |  | |||||||||
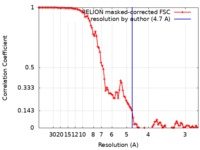
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Al-Otaibi NS / Farrell D | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: The cryo-EM structure of the bacterial flagellum cap complex suggests a molecular mechanism for filament elongation. Authors: Natalie S Al-Otaibi / Aidan J Taylor / Daniel P Farrell / Svetomir B Tzokov / Frank DiMaio / David J Kelly / Julien R C Bergeron /   Abstract: The bacterial flagellum is a remarkable molecular motor, whose primary function in bacteria is to facilitate motility through the rotation of a filament protruding from the bacterial cell. A cap ...The bacterial flagellum is a remarkable molecular motor, whose primary function in bacteria is to facilitate motility through the rotation of a filament protruding from the bacterial cell. A cap complex, consisting of an oligomer of the protein FliD, is localized at the tip of the flagellum, and is essential for filament assembly, as well as adherence to surfaces in some bacteria. However, the structure of the intact cap complex, and the molecular basis for its interaction with the filament, remains elusive. Here we report the cryo-EM structure of the Campylobacter jejuni cap complex, which reveals that FliD is pentameric, with the N-terminal region of the protomer forming an extensive set of contacts across several subunits, that contribute to FliD oligomerization. We also demonstrate that the native C. jejuni flagellum filament is 11-stranded, contrary to a previously published cryo-EM structure, and propose a molecular model for the filament-cap interaction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10210.map.gz emd_10210.map.gz | 73.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10210-v30.xml emd-10210-v30.xml emd-10210.xml emd-10210.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10210_fsc.xml emd_10210_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_10210.png emd_10210.png | 60.8 KB | ||
| Filedesc metadata |  emd-10210.cif.gz emd-10210.cif.gz | 5.5 KB | ||
| Others |  emd_10210_additional.map.gz emd_10210_additional.map.gz | 72.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10210 http://ftp.pdbj.org/pub/emdb/structures/EMD-10210 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10210 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10210 | HTTPS FTP |
-Related structure data
| Related structure data |  6sihMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10210.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10210.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FliDcj full model | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: FliDcj head domain
| File | emd_10210_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FliDcj head domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Decamer complex of FliD
| Entire | Name: Decamer complex of FliD |
|---|---|
| Components |
|
-Supramolecule #1: Decamer complex of FliD
| Supramolecule | Name: Decamer complex of FliD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Flagellar hook-associated protein 2
| Macromolecule | Name: Flagellar hook-associated protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.159305 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MAFGSLSSLG FGSGVLTQDT IDKLKEAEQK ARIDPYTKKI EENTTKQKDL TEIKTKLLSF QTAVSSLAD ATVFAKRKVV GSISDNPPAS LTVNSGVALQ SMNINVTQLA QKDVYQSKGL ANDSGFVNAN LTGTTDLTFF S NGKEYTVT ...String: MGSSHHHHHH SSGLVPRGSH MAFGSLSSLG FGSGVLTQDT IDKLKEAEQK ARIDPYTKKI EENTTKQKDL TEIKTKLLSF QTAVSSLAD ATVFAKRKVV GSISDNPPAS LTVNSGVALQ SMNINVTQLA QKDVYQSKGL ANDSGFVNAN LTGTTDLTFF S NGKEYTVT VDKNTTYRDL ADKINEASGG EIVAKIVNTG EKGTPYRLTL TSKETGEDSA ISFYAGKKDA QGQYQSDPEA EN IFSNLGW ELDKTTQTID PAKDKKGYGI KDASLHIQTA QNAEFTLDGI KMFRSSNTVT DLGVGMTLTL NKTGEINFDV QQD FEGVTK AMQDLVDAYN DLVTNLNAAT DYNSETGTKG TLQGISEVNS IRSSILADLF DSQVVDGTTE DANGNKVNTK VMLS MQDFG LSLNDAGTLS FDSSKFEQKV KEDPDSTESF FSNITKYEDI NHTGEVIKQG SLNQYLDSSG TGNKGLDFKP GDFTI VFNN QTYDLSKNSD GTNFKLTGKT EEELLQNLAN HINSKGIEGL KVKVESYDQN GVKGFKLNFS GDGSSDFSIK GNATIL QEL GLSDVNITSK PIEGKGIFSK LKATLQEMTG KDGSITKYDE SLTNDIKSLN TSKDSTQAMI DTRYDTMANQ WLQYESI LN KLNQQLNTVT NMINAANNSN N UniProtKB: Flagellar hook-associated protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1223 / Average exposure time: 10.0 sec. / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 36232 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)