[English] 日本語
 Yorodumi
Yorodumi- EMDB-10136: Double-stranded helical ESCRT-III filament formed from Snf7/Vps24... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10136 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical membrane bicelle | |||||||||||||||
 Map data Map data | Masked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical bicelle | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
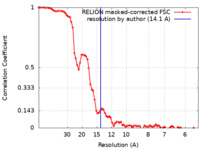
| Method | helical reconstruction / cryo EM / Resolution: 14.1 Å | |||||||||||||||
 Authors Authors | Frost A / Johnson I / Talledge N | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Anisotropic ESCRT-III architecture governs helical membrane tube formation. Authors: Joachim Moser von Filseck / Luca Barberi / Nathaniel Talledge / Isabel E Johnson / Adam Frost / Martin Lenz / Aurélien Roux /    Abstract: ESCRT-III proteins assemble into ubiquitous membrane-remodeling polymers during many cellular processes. Here we describe the structure of helical membrane tubes that are scaffolded by bundled ESCRT- ...ESCRT-III proteins assemble into ubiquitous membrane-remodeling polymers during many cellular processes. Here we describe the structure of helical membrane tubes that are scaffolded by bundled ESCRT-III filaments. Cryo-ET reveals how the shape of the helical membrane tube arises from the assembly of two distinct bundles of helical filaments that have the same helical path but bind the membrane with different interfaces. Higher-resolution cryo-EM of filaments bound to helical bicelles confirms that ESCRT-III filaments can interact with the membrane through a previously undescribed interface. Mathematical modeling demonstrates that the interface described above is key to the mechanical stability of helical membrane tubes and helps infer the rigidity of the described protein filaments. Altogether, our results suggest that the interactions between ESCRT-III filaments and the membrane could proceed through multiple interfaces, to provide assembly on membranes with various shapes, or adapt the orientation of the filaments towards the membrane during membrane remodeling. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10136.map.gz emd_10136.map.gz | 3.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10136-v30.xml emd-10136-v30.xml emd-10136.xml emd-10136.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10136_fsc.xml emd_10136_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10136.png emd_10136.png | 40.8 KB | ||
| Others |  emd_10136_additional.map.gz emd_10136_additional.map.gz emd_10136_additional_1.map.gz emd_10136_additional_1.map.gz emd_10136_half_map_1.map.gz emd_10136_half_map_1.map.gz emd_10136_half_map_2.map.gz emd_10136_half_map_2.map.gz | 36.1 MB 36.1 MB 31.1 MB 31.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10136 http://ftp.pdbj.org/pub/emdb/structures/EMD-10136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10136 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10136.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10136.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical bicelle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unmasked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2...
| File | emd_10136_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical bicelle | ||||||||||||
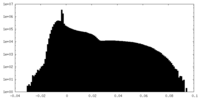
| Projections & Slices |
| ||||||||||||
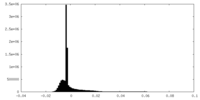
| Density Histograms |
-Additional map: Unmasked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2...
| File | emd_10136_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical bicelle | ||||||||||||
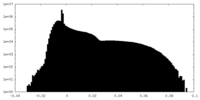
| Projections & Slices |
| ||||||||||||
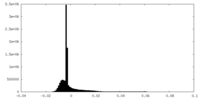
| Density Histograms |
-Half map: #1
| File | emd_10136_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10136_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Double-stranded helical ESCRT-III filament formed from Snf7/Vps24...
| Entire | Name: Double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical bicelle |
|---|---|
| Components |
|
-Supramolecule #1: Double-stranded helical ESCRT-III filament formed from Snf7/Vps24...
| Supramolecule | Name: Double-stranded helical ESCRT-III filament formed from Snf7/Vps24/Vps2 on a helical bicelle type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 30 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 67.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)