+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10058 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleosome-CHD4 complex structure (single CHD4 copy) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / ATPase / chromatin / nucleosome / CHD family / TRANSCRIPTION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcerebellar granule cell to Purkinje cell synapse / terminal button organization / NuRD complex / regulation of cell fate specification / NGF-stimulated transcription / regulation of stem cell differentiation / ATP-dependent chromatin remodeler activity / regulation of synapse assembly / RNA Polymerase I Transcription Initiation / site of DNA damage ...cerebellar granule cell to Purkinje cell synapse / terminal button organization / NuRD complex / regulation of cell fate specification / NGF-stimulated transcription / regulation of stem cell differentiation / ATP-dependent chromatin remodeler activity / regulation of synapse assembly / RNA Polymerase I Transcription Initiation / site of DNA damage / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / Regulation of TP53 Activity through Acetylation / Regulation of PTEN gene transcription / transcription coregulator binding / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / Regulation of endogenous retroelements by KRAB-ZFP proteins / HDACs deacetylate histones / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / helicase activity / double-strand break repair via homologous recombination / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / RNA polymerase II transcription regulator complex / histone deacetylase binding / structural constituent of chromatin / transcription corepressor activity / heterochromatin formation / nucleosome / nucleosome assembly / histone binding / Potential therapeutics for SARS / RNA polymerase II-specific DNA-binding transcription factor binding / chromosome, telomeric region / chromatin remodeling / protein heterodimerization activity / negative regulation of gene expression / negative regulation of DNA-templated transcription / chromatin binding / centrosome / positive regulation of DNA-templated transcription / chromatin / negative regulation of transcription by RNA polymerase II / ATP hydrolysis activity / protein-containing complex / DNA binding / zinc ion binding / nucleoplasm / ATP binding / membrane / nucleus / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
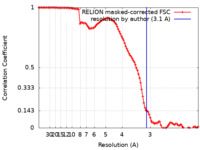
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Farnung L / Ochmann M | |||||||||||||||
| Funding support |  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. Authors: Lucas Farnung / Moritz Ochmann / Patrick Cramer /  Abstract: Chromatin remodeling plays important roles in gene regulation during development, differentiation and in disease. The chromatin remodeling enzyme CHD4 is a component of the NuRD and ChAHP complexes ...Chromatin remodeling plays important roles in gene regulation during development, differentiation and in disease. The chromatin remodeling enzyme CHD4 is a component of the NuRD and ChAHP complexes that are involved in gene repression. Here, we report the cryo-electron microscopy (cryo-EM) structure of CHD4 engaged with a nucleosome core particle in the presence of the non-hydrolysable ATP analogue AMP-PNP at an overall resolution of 3.1 Å. The ATPase motor of CHD4 binds and distorts nucleosomal DNA at superhelical location (SHL) +2, supporting the 'twist defect' model of chromatin remodeling. CHD4 does not induce unwrapping of terminal DNA, in contrast to its homologue Chd1, which functions in gene activation. Our structure also maps CHD4 mutations that are associated with human cancer or the intellectual disability disorder Sifrim-Hitz-Weiss syndrome. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10058.map.gz emd_10058.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10058-v30.xml emd-10058-v30.xml emd-10058.xml emd-10058.xml | 28.7 KB 28.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10058_fsc.xml emd_10058_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10058.png emd_10058.png | 40.8 KB | ||
| Masks |  emd_10058_msk_1.map emd_10058_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10058.cif.gz emd-10058.cif.gz | 7.8 KB | ||
| Others |  emd_10058_additional.map.gz emd_10058_additional.map.gz emd_10058_half_map_1.map.gz emd_10058_half_map_1.map.gz emd_10058_half_map_2.map.gz emd_10058_half_map_2.map.gz | 80.7 MB 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10058 http://ftp.pdbj.org/pub/emdb/structures/EMD-10058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10058 | HTTPS FTP |
-Related structure data
| Related structure data |  6ryrMC  6ryuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10411 (Title: Single Particle Cryo-EM Reconstructions of NCP-CHD4 complexes EMPIAR-10411 (Title: Single Particle Cryo-EM Reconstructions of NCP-CHD4 complexesData size: 721.2 Data #1: Unaligned multi-frame micrographs of NCP-CHD4 Dataset 1 [micrographs - multiframe] Data #2: Averaged and aligned micrographs Dataset 1 [micrographs - single frame] Data #3: Unaligned multi-frame micrographs of NCP-CHD4 Dataset 2 [micrographs - multiframe] Data #4: Averaged and aligned micrographs NCP-CHD4 Dataset 2 [micrographs - single frame] Data #5: Unaligned multi-frame micrographs of NCP-CHD4 Dataset 3 [micrographs - multiframe] Data #6: Averaged and aligned micrographs Dataset 3 [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10058.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10058.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
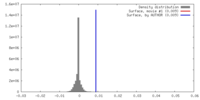
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10058_msk_1.map emd_10058_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
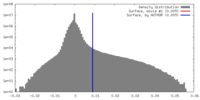
| Density Histograms |
-Additional map: #1
| File | emd_10058_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
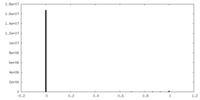
| Density Histograms |
-Half map: #1
| File | emd_10058_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10058_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Nucleosome-CHD4 complex
+Supramolecule #1: Nucleosome-CHD4 complex
+Supramolecule #2: Nucleosome
+Supramolecule #3: DNA
+Supramolecule #4: CHD4
+Macromolecule #1: Histone H3.2
+Macromolecule #2: Histone H4
+Macromolecule #3: Histone H2A type 1
+Macromolecule #4: Histone H2B 1.1
+Macromolecule #7: Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase...
+Macromolecule #5: DNA (149-MER)
+Macromolecule #6: DNA (149-MER)
+Macromolecule #8: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)