[English] 日本語
 Yorodumi
Yorodumi- EMDB-0205: Cryo-EM structure of the hydrazine dehydrogenase from Kuenia stut... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0205 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the hydrazine dehydrogenase from Kuenia stuttgartiensis in the octameric state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) | |||||||||
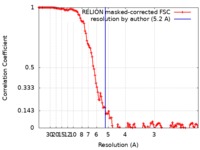
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Parey K / Barends TRM / Prinz S / Mohd A / Dietl A | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2019 Journal: Sci Adv / Year: 2019Title: A 192-heme electron transfer network in the hydrazine dehydrogenase complex. Authors: M Akram / A Dietl / U Mersdorf / S Prinz / W Maalcke / J Keltjens / C Ferousi / N M de Almeida / J Reimann / B Kartal / M S M Jetten / K Parey / T R M Barends /   Abstract: Anaerobic ammonium oxidation (anammox) is a major process in the biogeochemical nitrogen cycle in which nitrite and ammonium are converted to dinitrogen gas and water through the highly reactive ...Anaerobic ammonium oxidation (anammox) is a major process in the biogeochemical nitrogen cycle in which nitrite and ammonium are converted to dinitrogen gas and water through the highly reactive intermediate hydrazine. So far, it is unknown how anammox organisms convert the toxic hydrazine into nitrogen and harvest the extremely low potential electrons (-750 mV) released in this process. We report the crystal structure and cryo electron microscopy structures of the responsible enzyme, hydrazine dehydrogenase, which is a 1.7 MDa multiprotein complex containing an extended electron transfer network of 192 heme groups spanning the entire complex. This unique molecular arrangement suggests a way in which the protein stores and releases the electrons obtained from hydrazine conversion, the final step in the globally important anammox process. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0205.map.gz emd_0205.map.gz | 152.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0205-v30.xml emd-0205-v30.xml emd-0205.xml emd-0205.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0205_fsc.xml emd_0205_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_0205.png emd_0205.png | 245.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0205 http://ftp.pdbj.org/pub/emdb/structures/EMD-0205 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0205 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0205 | HTTPS FTP |
-Validation report
| Summary document |  emd_0205_validation.pdf.gz emd_0205_validation.pdf.gz | 327.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0205_full_validation.pdf.gz emd_0205_full_validation.pdf.gz | 326.7 KB | Display | |
| Data in XML |  emd_0205_validation.xml.gz emd_0205_validation.xml.gz | 13 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0205 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0205 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0205 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0205 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0205.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0205.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hydrazine dehydrogenase
| Entire | Name: Hydrazine dehydrogenase |
|---|---|
| Components |
|
-Supramolecule #1: Hydrazine dehydrogenase
| Supramolecule | Name: Hydrazine dehydrogenase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Molecular weight | Theoretical: 1.6 MDa |
-Macromolecule #1: Hydrazine dehydrogenase
| Macromolecule | Name: Hydrazine dehydrogenase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: hydrazine dehydrogenase |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Sequence | String: MRKFLKVTLA SALIGCGVIG TVSSLMVKEA KAVEIITHWV PHEVYGMPGE PDNSGKVFFS GLKAKYMGY PKDAQRSPYP GKYSKFWKTL PAYRYYIPDY MYNRDEVRPS NPIKGTFKLE Q CVACHSVM TPGIVRDYNK SAHSKAEPAP TGCDTCHGNN HQKLTMPSSK ...String: MRKFLKVTLA SALIGCGVIG TVSSLMVKEA KAVEIITHWV PHEVYGMPGE PDNSGKVFFS GLKAKYMGY PKDAQRSPYP GKYSKFWKTL PAYRYYIPDY MYNRDEVRPS NPIKGTFKLE Q CVACHSVM TPGIVRDYNK SAHSKAEPAP TGCDTCHGNN HQKLTMPSSK ACGTAECHET QY NEQGQGG IGSHASCSSF AQVECAWSIE RPPGDTAGCT FCHTSPEERC STCHQRHQFD PAV ARRSEQ CKTCHWGKDH RDWEAYDIGL HGTVYQVNKW DTEQFDFSKK LSDADYVGPT CQYC HMRGG HHNVQRASIV YTSMGMSMAD RGAPLWKEKR DRWVSICDDC HSPRFARENL QAMDE SVKD ASLKYRETFK VAEDLLIDGV LDPMPKDLCP DWSGQHIWSL KIGAYHDGEA YGGTTG ESG EFRMSNCTDV ERLCFESVGY FQTYIYKGMA HGSWNDATYS DGSFGMDRWL VNVKQNA SR ARRLAALEKK VGISWQPEQF WKTGEWLDQL TGPYIVKNHP GKTIFDLCPD PGWLDTHH A PAEEVEYIER KLKELGITAG SHSAHHHESG HDPAARSMKE H |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component - Concentration: 25.0 mM / Component - Formula: HEPES / Details: 25 mM HEPES/KOH, pH 7.5, 25 mM KCl |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | Glutaraldehyde cross-linked particles |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 452 / Average exposure time: 8.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 45872 / Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Cs: 2.0 mm / Nominal defocus max: -2.75 µm / Nominal defocus min: -0.75 µm / Nominal magnification: 200000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)