[English] 日本語
 Yorodumi
Yorodumi- SASDGL6: Ubiquitin activating enzyme 5 (UBA5_5) (Ubiquitin-like modifier-a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 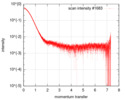 |
|---|---|
 Sample Sample | Ubiquitin activating enzyme 5 (UBA5_5)
|
| Function / homology |  Function and homology information Function and homology informationUFM1 activating enzyme activity / protein K69-linked ufmylation / protein ufmylation / megakaryocyte differentiation / regulation of type II interferon production / regulation of intracellular estrogen receptor signaling pathway / reticulophagy / neuromuscular process / response to endoplasmic reticulum stress / erythrocyte differentiation ...UFM1 activating enzyme activity / protein K69-linked ufmylation / protein ufmylation / megakaryocyte differentiation / regulation of type II interferon production / regulation of intracellular estrogen receptor signaling pathway / reticulophagy / neuromuscular process / response to endoplasmic reticulum stress / erythrocyte differentiation / Antigen processing: Ubiquitination & Proteasome degradation / intracellular membrane-bounded organelle / endoplasmic reticulum membrane / Golgi apparatus / protein homodimerization activity / zinc ion binding / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDGL6 SASDGL6 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Ubiquitin activating enzyme 5 (UBA5_5) / Specimen concentration: 4.38 mg/ml |
|---|---|
| Buffer | Name: 20 mM Tris, 150 mM NaCl, 2 mM DTT / pH: 7.5 |
| Entity #1904 | Name: UBA5 / Type: protein / Description: Ubiquitin-like modifier-activating enzyme 5 / Formula weight: 34.194 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: Q9GZZ9 Sequence: MGSSHHHHHH SSGLVPRGSM ALKRMGIVSD YEKIRTFAVA IVGVGGVGSV TAEMLTRCGI GKLLLFDYDK VELANMNRLF FQPHQAGLSK VQAAEHTLRN INPDVLFEVH NYNITTVENF QHFMDRISNG GLEEGKPVDL VLSCVDNFEA RMTINTACNE LGQTWMESGV ...Sequence: MGSSHHHHHH SSGLVPRGSM ALKRMGIVSD YEKIRTFAVA IVGVGGVGSV TAEMLTRCGI GKLLLFDYDK VELANMNRLF FQPHQAGLSK VQAAEHTLRN INPDVLFEVH NYNITTVENF QHFMDRISNG GLEEGKPVDL VLSCVDNFEA RMTINTACNE LGQTWMESGV SENAVSGHIQ LIIPGESACF ACAPPLVVAA NIDEKTLKRE GVCAASLPTT MGVVAGILVQ NVLKFLLNFG TVSFYLGYNA MQDFFPTMSM KPNPQCDDRN CRKQQEEYKK KVAALPKQEV IQEEEEIIHE DNEWGIELV |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.123982 Å / Dist. spec. to detc.: 3 mm / Type of source: X-ray synchrotron / Wavelength: 0.123982 Å / Dist. spec. to detc.: 3 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 6M | |||||||||||||||||||||||||||||||||
| Scan |
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller




