+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDG47 |
|---|---|
 Sample Sample | 2:4 heterohexamer of pUL7 and pUL51 from herpes simplex virus 1
|
| Function / homology | Herpesvirus UL51 / Herpesvirus UL51 protein / Herpesvirus UL7-like / Herpesvirus UL7 like / viral tegument / virion component / host cell Golgi apparatus / UL7 / UL51 Function and homology information Function and homology information |
| Biological species |   Human alphaherpesvirus 1 strain KOS Human alphaherpesvirus 1 strain KOS |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDG47 SASDG47 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
-Models
| Model #3903 | 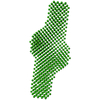 Type: dummy / Software: (SUPCOMB 23 (r10552)) / Radius of dummy atoms: 3.75 A / Symmetry: P2 Comment: Refined DAMMIN model of the HSV-1 pUL7:pUL51 2:4 heterohexamer Chi-square value: 0.910 / P-value: 0.805657 |
|---|---|
| Model #3904 |  Type: dummy / Software: (2.3i) / Radius of dummy atoms: 1.90 A / Symmetry: P2 Comment: Representative GASBOR model of the HSV-1 pUL7:pUL51 2:4 heterohexamer Chi-square value: 1.00 / P-value: 0.363345 |
- Sample
Sample
 Sample Sample | Name: 2:4 heterohexamer of pUL7 and pUL51 from herpes simplex virus 1 Specimen concentration: 8 mg/ml / Entity id: 1941 / 1942 |
|---|---|
| Buffer | Name: 20 mM HEPES, 200 mM NaCl, 3% (v/v) glycerol, 1 mM DTT / pH: 7.5 |
| Entity #1941 | Name: pUL7 / Type: protein / Description: Tegument protein UL7 / Formula weight: 33.907 / Num. of mol.: 2 / Source: Human alphaherpesvirus 1 strain KOS / References: UniProt: A0A110B4Q7 Sequence: MAAATADDEG SAATILKQAI AGDRSLVEAA EAISQQTLLR LACEVRQVGD RQPRFTATSI ARVDVAPGCR LRFVLDGSPE DAYVTSEDYF KRCCGQSSYR GFAVAVLTAN EDHVHSLAVP PLVLLHRFSL FNPRDLLDFE LACLLMYLEN CPRSHATPST FAKVLAWLGV ...Sequence: MAAATADDEG SAATILKQAI AGDRSLVEAA EAISQQTLLR LACEVRQVGD RQPRFTATSI ARVDVAPGCR LRFVLDGSPE DAYVTSEDYF KRCCGQSSYR GFAVAVLTAN EDHVHSLAVP PLVLLHRFSL FNPRDLLDFE LACLLMYLEN CPRSHATPST FAKVLAWLGV AGRRTSPFER VRCLFLRSCH WVLNTLMFMV HVKPFDDEFV LPHWYMARYL LANNPPPVLS ALFCATPTSS SFRLPGPPPR SDCVAYNPAG IMGSCWASEE VRAPLVYWWL SETPKRQTSS LFYQFCGSLE VLFQ |
| Entity #1942 | Name: pUL51 / Type: protein / Description: Tegument protein UL51 / Formula weight: 25.448 / Num. of mol.: 4 / Source: Human alphaherpesvirus 1 strain KOS / References: UniProt: D3YPL0 Sequence: MASLLGAISG WGARPEEQYE MIRAAVPPSE AEPRLQEALV VVNALLPAPI TLDDALGSLD DTRRLVKARA LARTYHACMV NLERLARHHP GLEAPTIDGA VAAHQDKMRR LADTCMATIL QMYMSVGAAD KSADVLVSQA IRSMAESDVV MEDVAIAERA LGLSAFGVAG ...Sequence: MASLLGAISG WGARPEEQYE MIRAAVPPSE AEPRLQEALV VVNALLPAPI TLDDALGSLD DTRRLVKARA LARTYHACMV NLERLARHHP GLEAPTIDGA VAAHQDKMRR LADTCMATIL QMYMSVGAAD KSADVLVSQA IRSMAESDVV MEDVAIAERA LGLSAFGVAG GTRSGGLGVT EAPSLGHPHT PPPEVTLAPA ARNGDALPDP KPESCPRVSV PRPTASPTAP RPGPSRAAPC VLGQ |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 6M | ||||||||||||||||||||||||||||||
| Scan | Measurement date: May 16, 2019 / Cell temperature: 20.2 °C / Exposure time: 0.995 sec. / Number of frames: 48 / Unit: 1/nm /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result | Comments: The quoted experimental molecular mass, that corresponds to a 2:4 pUL7:pUL51 heterohexamer, was determined from SEC-MALLS-RI measurements performed in parallel to the SEC-SAXS. The SEC-SAXS ...Comments: The quoted experimental molecular mass, that corresponds to a 2:4 pUL7:pUL51 heterohexamer, was determined from SEC-MALLS-RI measurements performed in parallel to the SEC-SAXS. The SEC-SAXS data, Rg correlations through the dimer elution peak as well as the the SEC-MALLS-RI data are made available in the full-entry zip archive. The GASBORMX model displayed in this entry represents a 100 % volume fraction of the heterohexamer in solution that has been separated from a 1:2 pUL7:pUL51 heterotrimer (refer to SASBDB entry SASDG57).
|
 Movie
Movie Controller
Controller



