+ Open data
Open data
- Basic information
Basic information
| Entry | 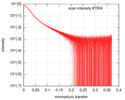 |
|---|---|
 Sample Sample | Apolipoprotein E2 (ApoE2) bound to 330 µg/mL heparin
|
| Function / homology |  Function and homology information Function and homology informationlipid transport involved in lipid storage / intermediate-density lipoprotein particle clearance / positive regulation of lipid transport across blood-brain barrier / regulation of cellular response to very-low-density lipoprotein particle stimulus / metal chelating activity / triglyceride-rich lipoprotein particle clearance / discoidal high-density lipoprotein particle / lipoprotein particle / negative regulation of triglyceride metabolic process / maintenance of location in cell ...lipid transport involved in lipid storage / intermediate-density lipoprotein particle clearance / positive regulation of lipid transport across blood-brain barrier / regulation of cellular response to very-low-density lipoprotein particle stimulus / metal chelating activity / triglyceride-rich lipoprotein particle clearance / discoidal high-density lipoprotein particle / lipoprotein particle / negative regulation of triglyceride metabolic process / maintenance of location in cell / regulation of amyloid-beta clearance / negative regulation of cholesterol biosynthetic process / positive regulation of lipoprotein transport / Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors / chylomicron remnant clearance / chylomicron remnant / intermediate-density lipoprotein particle / acylglycerol homeostasis / NMDA glutamate receptor clustering / very-low-density lipoprotein particle remodeling / Chylomicron clearance / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux / Chylomicron remodeling / lipid transporter activity / positive regulation of low-density lipoprotein particle receptor catabolic process / cellular response to lipoprotein particle stimulus / regulation of protein metabolic process / very-low-density lipoprotein particle clearance / Chylomicron assembly / response to caloric restriction / high-density lipoprotein particle clearance / phospholipid efflux / chylomicron / very-low-density lipoprotein particle receptor binding / regulation of amyloid fibril formation / lipoprotein catabolic process / AMPA glutamate receptor clustering / high-density lipoprotein particle remodeling / positive regulation of cholesterol metabolic process / melanosome organization / regulation of behavioral fear response / multivesicular body, internal vesicle / reverse cholesterol transport / positive regulation of amyloid-beta clearance / high-density lipoprotein particle assembly / host-mediated activation of viral process / low-density lipoprotein particle / lipoprotein biosynthetic process / cholesterol transfer activity / high-density lipoprotein particle / protein import / very-low-density lipoprotein particle / cholesterol catabolic process / heparan sulfate proteoglycan binding / low-density lipoprotein particle remodeling / amyloid precursor protein metabolic process / negative regulation of amyloid fibril formation / positive regulation of membrane protein ectodomain proteolysis / regulation of Cdc42 protein signal transduction / synaptic transmission, cholinergic / regulation of amyloid precursor protein catabolic process / HDL remodeling / negative regulation of endothelial cell migration / cholesterol efflux / regulation of cholesterol metabolic process / artery morphogenesis / negative regulation of protein metabolic process / regulation of axon extension / Scavenging by Class A Receptors / triglyceride homeostasis / triglyceride metabolic process / low-density lipoprotein particle receptor binding / positive regulation of amyloid fibril formation / regulation of innate immune response / virion assembly / negative regulation of endothelial cell proliferation / positive regulation of dendritic spine development / antioxidant activity / response to dietary excess / negative regulation of MAP kinase activity / lipoprotein particle binding / negative regulation of amyloid-beta formation / locomotory exploration behavior / negative regulation of long-term synaptic potentiation / negative regulation of blood vessel endothelial cell migration / negative regulation of platelet activation / negative regulation of blood coagulation / positive regulation of dendritic spine maintenance / positive regulation of endocytosis / regulation of neuronal synaptic plasticity / positive regulation of cholesterol efflux / negative regulation of protein secretion / fatty acid homeostasis / regulation of protein-containing complex assembly / long-term memory / long-chain fatty acid transport / synaptic cleft / intracellular transport / Nuclear signaling by ERBB4 Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDFZ8 SASDFZ8 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Apolipoprotein E2 (ApoE2) bound to 330 µg/mL heparin / Specimen concentration: 4 mg/ml / Entity id: 1086 / 1633 |
|---|---|
| Buffer | Name: 20 mM HEPES, 300 mM NaCl, 1 mM TCEP / pH: 8 |
| Entity #1086 | Name: ApoE2 / Type: protein / Description: Apolipoprotein E2 / Formula weight: 34.606 / Num. of mol.: 4 / Source: Homo sapiens / References: UniProt: P02649 Sequence: GPHMKVEQAV ETEPEPELRQ QTEWQSGQRW ELALGRFWDY LRWVQTLSEQ VQEELLSSQV TQELRALMDE TMKELKAYKS ELEEQLTPVA EETRARLSKE LQAAQARLGA DMEDVCGRLV QYRGEVQAML GQSTEELRVR LASHLRKLRK RLLRDADDLQ KCLAVYQAGA ...Sequence: GPHMKVEQAV ETEPEPELRQ QTEWQSGQRW ELALGRFWDY LRWVQTLSEQ VQEELLSSQV TQELRALMDE TMKELKAYKS ELEEQLTPVA EETRARLSKE LQAAQARLGA DMEDVCGRLV QYRGEVQAML GQSTEELRVR LASHLRKLRK RLLRDADDLQ KCLAVYQAGA REGAERGLSA IRERLGPLVE QGRVRAATVG SLAGQPLQER AQAWGERLRA RMEEMGSRTR DRLDEVKEQV AEVRAKLEEQ AQQIRLQAEA FQARLKSWFE PLVEDMQRQW AGLVEKVQAA VGTSAAPVPS DNH |
| Entity #1633 | Type: other / Description: Heparin / Formula weight: 15 / Num. of mol.: 1 Sequence: (C12H19NO20S3)n |
-Experimental information
| Beam | Instrument name: Diamond Light Source B21 / City: Didcot / 国: UK  / Shape: 1 x 5 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 4.036 mm / Shape: 1 x 5 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 4.036 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||
| Scan | Measurement date: May 3, 2018 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 1 sec. / Number of frames: 28 / Unit: 1/A /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result | Comments: ApoE2 + 330 µg/mL heparin. Molecular weight estimates are inaccurate due to heparin heterogeneity.
|
 Movie
Movie Controller
Controller















