+ Open data
Open data
- Basic information
Basic information
| Entry | 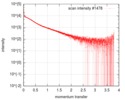 |
|---|---|
 Sample Sample | Delta subunit of RNA polymerase, RNAP (B. subtilis), 400 mM NaCl
|
| Function / homology |  Function and homology information Function and homology informationDNA-directed RNA polymerase complex / DNA-directed RNA polymerase activity / DNA-templated transcription / regulation of DNA-templated transcription Similarity search - Function |
| Biological species |  |
 Citation Citation |  Journal: J Am Chem Soc / Year: 2019 Journal: J Am Chem Soc / Year: 2019Title: Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase. Authors: Vojtěch Kubáň / Pavel Srb / Hana Štégnerová / Petr Padrta / Milan Zachrdla / Zuzana Jaseňáková / Hana Šanderová / Dragana Vítovská / Libor Krásný / Tomáš Koval' / Jan ...Authors: Vojtěch Kubáň / Pavel Srb / Hana Štégnerová / Petr Padrta / Milan Zachrdla / Zuzana Jaseňáková / Hana Šanderová / Dragana Vítovská / Libor Krásný / Tomáš Koval' / Jan Dohnálek / Joanna Ziemska-Legiecka / Marcin Grynberg / Patryk Jarnot / Aleksandra Gruca / Malene Ringkjøbing Jensen / Martin Blackledge / Lukáš Žídek /    Abstract: Electrostatic interactions play important roles in the functional mechanisms exploited by intrinsically disordered proteins (IDPs). The atomic resolution description of long-range and local ...Electrostatic interactions play important roles in the functional mechanisms exploited by intrinsically disordered proteins (IDPs). The atomic resolution description of long-range and local structural propensities that can both be crucial for the function of highly charged IDPs presents significant experimental challenges. Here, we investigate the conformational behavior of the δ subunit of RNA polymerase from whose unfolded domain is highly charged, with 7 positively charged amino acids followed by 51 acidic amino acids. Using a specifically designed analytical strategy, we identify transient contacts between the two regions using a combination of NMR paramagnetic relaxation enhancements, residual dipolar couplings (RDCs), chemical shifts, and small-angle scattering. This strategy allows the resolution of long-range and local ensemble averaged structural contributions to the experimental RDCs, and reveals that the negatively charged segment folds back onto the positively charged strand, compacting the conformational sampling of the protein while remaining highly flexible in solution. Mutation of the positively charged region abrogates the long-range contact, leaving the disordered domain in an extended conformation, possibly due to local repulsion of like-charges along the chain. Remarkably, in vitro studies show that this mutation also has a significant effect on transcription activity, and results in diminished cell fitness of the mutated bacteria in vivo. This study highlights the importance of accurately describing electrostatic interactions for understanding the functional mechanisms of IDPs. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Delta subunit of RNA polymerase, RNAP (B. subtilis), 400 mM NaCl Specimen concentration: 3.6 mg/ml |
|---|---|
| Buffer | Name: 20 mM Phosphate buffer, 400 mM NaCl, 0.05% NaN3 / pH: 6.6 |
| Entity #1736 | Name: RpoE / Type: protein / Description: DNA-directed RNA polymerase subunit delta / Formula weight: 20.399 / Num. of mol.: 1 / Source: Bacillus subtilis (strain 168) / References: UniProt: P12464 Sequence: MGIKQYSQEE LKEMALVEIA HELFEEHKKP VPFQELLNEI ASLLGVKKEE LGDRIAQFYT DLNIDGRFLA LSDQTWGLRS WYPYDQLDEE TQPTVKAKKK KAKKAVEEDL DLDEFEEIDE DDLDLDEVEE ELDLEADDFD EEDLDEDDDD LEIEEDIIDE DDEDYDDEEE EIK |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||||||||
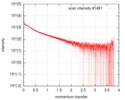
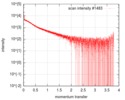
| Scan | Measurement date: Oct 3, 2016 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 0.05 sec. / Number of frames: 20 / Unit: 1/nm /
| |||||||||||||||||||||||||||
| Result | Type of curve: single_conc /
|
 Movie
Movie Controller
Controller



 SASDF78
SASDF78