[English] 日本語
 Yorodumi
Yorodumi- SASDEX4: Herpes simplex virus 1 tegument protein UL11 (Cytoplasmic envelop... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 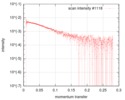 |
|---|---|
 Sample Sample | Herpes simplex virus 1 tegument protein UL11
|
| Function / homology |  Function and homology information Function and homology informationviral budding from Golgi membrane / viral tegument / host cell Golgi membrane / anatomical structure morphogenesis / host cell plasma membrane / virion membrane / membrane Similarity search - Function |
| Biological species |  Human herpesvirus 1 (strain 17) Human herpesvirus 1 (strain 17) |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDEX4 SASDEX4 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Herpes simplex virus 1 tegument protein UL11 / Specimen concentration: 18 mg/ml |
|---|---|
| Buffer | Name: 50 mM HEPES, 100 mM NaCl, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Entity #1315 | Name: UL11 / Type: protein / Description: Cytoplasmic envelopment protein 3 / Formula weight: 11.557 / Num. of mol.: 1 / Source: Human herpesvirus 1 (strain 17) / References: UniProt: P04289 Sequence: MGLSFSGTRP CCCRNNVLIT DDGEVVSLTA HDFDVVDIES EEEGNFYVPP DMRGVTRAPG RQRLRSSDPP SRHTHRRTPG GACPATQFPP PMSDSEWSHP QFEK |
-Experimental information
| Beam | Instrument name: Cornell High Energy Synchrotron Source (CHESS) G1 City: Ithaca, NY / 国: USA  / Type of source: X-ray synchrotron / Wavelength: 0.124612 Å / Dist. spec. to detc.: 1.52109 mm / Type of source: X-ray synchrotron / Wavelength: 0.124612 Å / Dist. spec. to detc.: 1.52109 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 200K / Type: Pilatus / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


