[English] 日本語
 Yorodumi
Yorodumi- SASDEV4: ACA8 protein (apo) in stealth nanodisc (Membrane scaffold protein... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 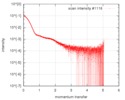 |
|---|---|
 Sample Sample | ACA8 protein (apo) in stealth nanodisc
|
| Function / homology |  Function and homology information Function and homology informationP-type Ca2+ transporter / P-type calcium transporter activity / plasmodesma / response to nematode / plastid / calmodulin binding / ATP hydrolysis activity / ATP binding / metal ion binding / plasma membrane Similarity search - Function |
| Biological species | Escherichia coli. (AL95 strain) |
 Citation Citation |  Date: 2018 Dec Date: 2018 DecTitle: Structural basis for activation of plasma-membrane Ca2+-ATPase by calmodulin Authors: Nitsche J / Josts I / Heidemann J / Mertens H / Maric S / Moulin M / Haertlein M / Busch S / Forsyth V / Svergun D / Uetrecht C |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDEV4 SASDEV4 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: ACA8 protein (apo) in stealth nanodisc / Contrast: 1.808 / Specific vol: 0.816 / Specimen concentration: 1.00-8.00 / Concentration method: A280 / Entity id: 1017 / 1018 / 1312 |
|---|---|
| Buffer | Name: 30 mM Tris, 150 mM NaCl, 1mM MgCl2, 1 mM CaCl2 / pH: 7.5 |
| Entity #1017 | Name: d75-msp1d1 / Type: protein Description: Membrane scaffold protein 1D1 (deuterated, 75%) Formula weight: 24.661 / Num. of mol.: 2 Sequence: GHHHHHHHDY DIPTTENLYF QGSTFSKLRE QLGPVTQEFW DNLEKETEGL RQEMSKDLEE VKAKVQPYLD DFQKKWQEEM ELYRQKVEPL RAELQEGARQ KLHELQEKLS PLGEEMRDRA RAHVDALRTH LAPYSDELRQ RLAARLEALK ENGGARLAEY HAKATEHLST ...Sequence: GHHHHHHHDY DIPTTENLYF QGSTFSKLRE QLGPVTQEFW DNLEKETEGL RQEMSKDLEE VKAKVQPYLD DFQKKWQEEM ELYRQKVEPL RAELQEGARQ KLHELQEKLS PLGEEMRDRA RAHVDALRTH LAPYSDELRQ RLAARLEALK ENGGARLAEY HAKATEHLST LSEKAKPALE DLRQGLLPVL ESFKVSFLSA LEEYTKKLNT Q |
| Entity #1018 | Name: d(78,92)-POPC / Type: other Description: 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl) Formula weight: 0.76 / Source: Escherichia coli. (AL95 strain) Sequence: C42H82NO8P |
| Entity #1312 | Name: ACA8 / Type: protein Description: Calcium-transporting ATPase 8, plasma membrane-type Formula weight: 118.209 / Num. of mol.: 1 / Source: Arabidopsis thaliana / References: UniProt: Q9LF79 Sequence: HHHHHHHHEN LYFQGSMTSL LKSSPGRRRG GDVESGKSEH ADSDSDTFYI PSKNASIERL QQWRKAALVL NASRRFRYTL DLKKEQETRE MRQKIRSHAH ALLAANRFMD MGRESGVEKT TGPATPAGDF GITPEQLVIM SKDHNSGALE QYGGTQGLAN LLKTNPEKGI ...Sequence: HHHHHHHHEN LYFQGSMTSL LKSSPGRRRG GDVESGKSEH ADSDSDTFYI PSKNASIERL QQWRKAALVL NASRRFRYTL DLKKEQETRE MRQKIRSHAH ALLAANRFMD MGRESGVEKT TGPATPAGDF GITPEQLVIM SKDHNSGALE QYGGTQGLAN LLKTNPEKGI SGDDDDLLKR KTIYGSNTYP RKKGKGFLRF LWDACHDLTL IILMVAAVAS LALGIKTEGI KEGWYDGGSI AFAVILVIVV TAVSDYKQSL QFQNLNDEKR NIHLEVLRGG RRVEISIYDI VVGDVIPLNI GNQVPADGVL ISGHSLALDE SSMTGESKIV NKDANKDPFL MSGCKVADGN GSMLVTGVGV NTEWGLLMAS ISEDNGEETP LQVRLNGVAT FIGSIGLAVA AAVLVILLTR YFTGHTKDNN GGPQFVKGKT KVGHVIDDVV KVLTVAVTIV VVAVPEGLPL AVTLTLAYSM RKMMADKALV RRLSACETMG SATTICSDKT GTLTLNQMTV VESYAGGKKT DTEQLPATIT SLVVEGISQN TTGSIFVPEG GGDLEYSGSP TEKAILGWGV KLGMNFETAR SQSSILHAFP FNSEKKRGGV AVKTADGEVH VHWKGASEIV LASCRSYIDE DGNVAPMTDD KASFFKNGIN DMAGRTLRCV ALAFRTYEAE KVPTGEELSK WVLPEDDLIL LAIVGIKDPC RPGVKDSVVL CQNAGVKVRM VTGDNVQTAR AIALECGILS SDADLSEPTL IEGKSFREMT DAERDKISDK ISVMGRSSPN DKLLLVQSLR RQGHVVAVTG DGTNDAPALH EADIGLAMGI AGTEVAKESS DIIILDDNFA SVVKVVRWGR SVYANIQKFI QFQLTVNVAA LVINVVAAIS SGDVPLTAVQ LLWVNLIMDT LGALALATEP PTDHLMGRPP VGRKEPLITN IMWRNLLIQA IYQVSVLLTL NFRGISILGL EHEVHEHATR VKNTIIFNAF VLCQAFNEFN ARKPDEKNIF KGVIKNRLFM GIIVITLVLQ VIIVEFLGKF ASTTKLNWKQ WLICVGIGVI SWPLALVGKF IPVPAAPISN KLKVLKFWGK KKNSSGEGSL |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.3 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.3 mm | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||||||||||||||
| Result | Comments: 'ACA8 protein (apo) encapsulated in a partially deuterated
|
 Movie
Movie Controller
Controller





