+ Open data
Open data
- Basic information
Basic information
| Entry | 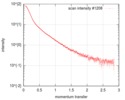 |
|---|---|
 Sample Sample | KRAB-associated protein 1 (KAP1); TRIM28, amino acids 23-812
|
| Function / homology |  Function and homology information Function and homology informationconvergent extension involved in axis elongation / Krueppel-associated box domain binding / embryonic placenta morphogenesis / negative regulation of single stranded viral RNA replication via double stranded DNA intermediate / suppression of viral release by host / chromo shadow domain binding / genomic imprinting / Generic Transcription Pathway / SUMO transferase activity / DNA methylation-dependent constitutive heterochromatin formation ...convergent extension involved in axis elongation / Krueppel-associated box domain binding / embryonic placenta morphogenesis / negative regulation of single stranded viral RNA replication via double stranded DNA intermediate / suppression of viral release by host / chromo shadow domain binding / genomic imprinting / Generic Transcription Pathway / SUMO transferase activity / DNA methylation-dependent constitutive heterochromatin formation / protein sumoylation / epithelial to mesenchymal transition / heterochromatin / embryo implantation / SUMOylation of transcription cofactors / positive regulation of DNA repair / Regulation of endogenous retroelements by KRAB-ZFP proteins / promoter-specific chromatin binding / euchromatin / RING-type E3 ubiquitin transferase / positive regulation of protein import into nucleus / RNA polymerase II transcription regulator complex / HCMV Early Events / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / transcription corepressor activity / chromatin organization / proteasome-mediated ubiquitin-dependent protein catabolic process / transcription coactivator activity / protein kinase activity / innate immune response / DNA repair / negative regulation of DNA-templated transcription / ubiquitin protein ligase binding / chromatin binding / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / RNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Life Sci Alliance / Year: 2019 Journal: Life Sci Alliance / Year: 2019Title: KAP1 is an antiparallel dimer with a functional asymmetry. Authors: Giulia Fonti / Maria J Marcaida / Louise C Bryan / Sylvain Träger / Alexandra S Kalantzi / Pierre-Yves Jl Helleboid / Davide Demurtas / Mark D Tully / Sergei Grudinin / Didier Trono / Beat ...Authors: Giulia Fonti / Maria J Marcaida / Louise C Bryan / Sylvain Träger / Alexandra S Kalantzi / Pierre-Yves Jl Helleboid / Davide Demurtas / Mark D Tully / Sergei Grudinin / Didier Trono / Beat Fierz / Matteo Dal Peraro /   Abstract: KAP1 (KRAB domain-associated protein 1) plays a fundamental role in regulating gene expression in mammalian cells by recruiting different transcription factors and altering the chromatin state. In ...KAP1 (KRAB domain-associated protein 1) plays a fundamental role in regulating gene expression in mammalian cells by recruiting different transcription factors and altering the chromatin state. In doing so, KAP1 acts both as a platform for macromolecular interactions and as an E3 small ubiquitin modifier ligase. This work sheds light on the overall organization of the full-length protein combining solution scattering data, integrative modeling, and single-molecule experiments. We show that KAP1 is an elongated antiparallel dimer with an asymmetry at the C-terminal domains. This conformation is consistent with the finding that the Really Interesting New Gene (RING) domain contributes to KAP1 auto-SUMOylation. Importantly, this intrinsic asymmetry has key functional implications for the KAP1 network of interactions, as the heterochromatin protein 1 (HP1) occupies only one of the two putative HP1 binding sites on the KAP1 dimer, resulting in an unexpected stoichiometry, even in the context of chromatin fibers. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDER7 SASDER7 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: KRAB-associated protein 1 (KAP1); TRIM28, amino acids 23-812 Specimen concentration: 12 mg/ml |
|---|---|
| Buffer | Name: 20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP / pH: 7.5 |
| Entity #1378 | Name: KAP1, residues23-812 / Type: protein Description: Transcription intermediary factor 1-beta, TIF1b, KAP1, TRIM28; amino acids 23-812 Formula weight: 87.513 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: Q13263 Sequence: MGSSHHHHHH SQDPNSSSEN LYFQGEGSAG GEKRSTAPSA AASASASAAA SSPAGGGAEA LELLEHCGVC RERLRPEREP RLLPCLHSAC SACLGPAAPA AANSSGDGGA AGDGTVVDCP VCKQQCFSKD IVENYFMRDS GSKAATDAQD ANQCCTSCED NAPATSYCVE ...Sequence: MGSSHHHHHH SQDPNSSSEN LYFQGEGSAG GEKRSTAPSA AASASASAAA SSPAGGGAEA LELLEHCGVC RERLRPEREP RLLPCLHSAC SACLGPAAPA AANSSGDGGA AGDGTVVDCP VCKQQCFSKD IVENYFMRDS GSKAATDAQD ANQCCTSCED NAPATSYCVE CSEPLCETCV EAHQRVKYTK DHTVRSTGPA KSRDGERTVY CNVHKHEPLV LFCESCDTLT CRDCQLNAHK DHQYQFLEDA VRNQRKLLAS LVKRLGDKHA TLQKSTKEVR SSIRQVSDVQ KRVQVDVKMA ILQIMKELNK RGRVLVNDAQ KVTEGQQERL ERQHWTMTKI QKHQEHILRF ASWALESDNN TALLLSKKLI YFQLHRALKM IVDPVEPHGE MKFQWDLNAW TKSAEAFGKI VAERPGTNST GPAPMAPPRA PGPLSKQGSG SSQPMEVQEG YGFGSGDDPY SSAEPHVSGV KRSRSGEGEV SGLMRKVPRV SLERLDLDLT ADSQPPVFKV FPGSTTEDYN LIVIERGAAA AATGQPGTAP AGTPGAPPLA GMAIVKEEET EAAIGAPPTA TEGPETKPVL MALAEGPGAE GPRLASPSGS TSSGLEVVAP EGTSAPGGGP GTLDDSATIC RVCQKPGDLV MCNQCEFCFH LDCHLPALQD VPGEEWSCSL CHVLPDLKEE DGSLSLDGAD STGVVAKLSP ANQRKCERVL LALFCHEPCR PLHQLATDST FSLDQPGGTL DLTLIRARLQ EKLSPPYSSP QEFAQDVGRM FKQFNKLTED KADVQSIIGL QRFFETRMNE AFGDTKFSAV LVEP |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.867 mm / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.867 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
| Scan | Measurement date: Jul 6, 2017 / Cell temperature: 20 °C / Exposure time: 1 sec. / Number of frames: 3000 / Unit: 1/nm /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller







