[English] 日本語
 Yorodumi
Yorodumi- SASDEM7: Human glycine decarboxylase with glycine (Glycine decarboxylase, GLDC) -
+ Open data
Open data
- Basic information
Basic information
| Entry | 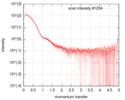 |
|---|---|
 Sample Sample | Human glycine decarboxylase with glycine
|
| Function / homology |  Function and homology information Function and homology informationresponse to methylamine / response to lipoic acid / pyridoxal binding / glycine dehydrogenase (aminomethyl-transferring) / glycine dehydrogenase (decarboxylating) activity / glycine catabolic process / Glycine degradation / glycine decarboxylation via glycine cleavage system / glycine cleavage complex / glycine binding ...response to methylamine / response to lipoic acid / pyridoxal binding / glycine dehydrogenase (aminomethyl-transferring) / glycine dehydrogenase (decarboxylating) activity / glycine catabolic process / Glycine degradation / glycine decarboxylation via glycine cleavage system / glycine cleavage complex / glycine binding / cellular response to leukemia inhibitory factor / pyridoxal phosphate binding / electron transfer activity / lyase activity / mitochondrial matrix / protein homodimerization activity / mitochondrion / nucleoplasm / plasma membrane Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDEM7 SASDEM7 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Human glycine decarboxylase with glycine / Specimen concentration: 5 mg/ml |
|---|---|
| Buffer | Name: 10 mM TRIS pH 7.5, 100 mM NaCl, 1 mM DTT , 100 mM glycine pH: 7.5 |
| Entity #1368 | Name: GLDC / Type: protein / Description: Glycine decarboxylase / Formula weight: 109.367 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: P23378 Sequence: MAAAGASRLL ERLLPRHDDF ARRHIGPGDK DQREMLQTLG LASIDELIEK TVPANIRLKR PLKMEDPVCE NEILATLHAI SSKNQIWRSY IGMGYYNCSV PQTILRNLLE NSGWITQYTP YQPEVSQGRL ESLLNYQTMV CDITGLDMAN ASLLDEGTAA AEALQLCYRH ...Sequence: MAAAGASRLL ERLLPRHDDF ARRHIGPGDK DQREMLQTLG LASIDELIEK TVPANIRLKR PLKMEDPVCE NEILATLHAI SSKNQIWRSY IGMGYYNCSV PQTILRNLLE NSGWITQYTP YQPEVSQGRL ESLLNYQTMV CDITGLDMAN ASLLDEGTAA AEALQLCYRH NKRRKFLVDP RCHPQTIAVV QTRAKYTGVL TELKLPCEMD FSGKDVSGVL FQYPDTEGKV EDFTELVERA HQSGSLACCA TDLLALCILR PPGEFGVDIA LGSSQRFGVP LGYGGPHAAF FAVRESLVRM MPGRMVGVTR DATGKEVYRL ALQTREQHIR RDKATSNICT AQALLANMAA MFAIYHGSHG LEHIARRVHN ATLILSEGLK RAGHQLQHDL FFDTLKIQCG CSVKEVLGRA AQRQINFRLF EDGTLGISLD ETVNEKDLDD LLWIFGCESS AELVAESMGE ECRGIPGSVF KRTSPFLTHQ VFNSYHSETN IVRYMKKLEN KDISLVHSMI PLGSCTMKLN SSSELAPITW KEFANIHPFV PLDQAQGYQQ LFRELEKDLC ELTGYDQVCF QPNSGAQGEY AGLATIRAYL NQKGEGHRTV CLIPKSAHGT NPASAHMAGM KIQPVEVDKY GNIDAVHLKA MVDKHKENLA AIMITYPSTN GVFEENISDV CDLIHQHGGQ VYLDGANMNA QVGICRPGDF GSDVSHLNLH KTFCIPHGGG GPGMGPIGVK KHLAPFLPNH PVISLKRNED ACPVGTVSAA PWGSSSILPI SWAYIKMMGG KGLKQATETA ILNANYMAKR LETHYRILFR GARGYVGHEF ILDTRPFKKS ANIEAVDVAK RLQDYGFHAP TMSWPVAGTL MVEPTESEDK AELDRFCDAM ISIRQEIADI EEGRIDPRVN PLKMSPHSLT CVTSSHWDRP YSREVAAFPL PFVKPENKFW PTIARIDDIY GDQHLVCTCP PMEVYESPFS EQKRASSGHH HHHH |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.864 mm / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.864 mm | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||
| Result | Type of curve: single_conc /
|
 Movie
Movie Controller
Controller


