+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Cell wall synthesis protein Wag31: Polymer tbWag31 T73A
|
| Function / homology |  Function and homology information Function and homology informationpeptidoglycan-based cell wall biogenesis / regulation of growth rate / cell pole / peptidoglycan-based cell wall / regulation of cell shape / protein stabilization / cell division / plasma membrane / cytoplasm Similarity search - Function |
| Biological species |  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) |
 Citation Citation |  Date: 2018 Aug 18 Date: 2018 Aug 18Title: Higher order assembling of the mycobacterial essential polar growth factor DivIVA/Wag31 Authors: Choukate K / Gupta A / Basu B / Virk K / Ganguli M |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDEC2 SASDEC2 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
-Models
- Sample
Sample
 Sample Sample | Name: Cell wall synthesis protein Wag31: Polymer tbWag31 T73A Specimen concentration: 2.7 mg/ml |
|---|---|
| Buffer | Name: 50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME) pH: 7.5 |
| Entity #1213 | Type: protein / Description: Cell wall synthesis protein Wag31 / Formula weight: 29.922 Source: Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) References: UniProt: P9WMU1 Sequence: MHHHHHHENL YFQGPLTPAD VHNVAFSKPP IGKRGYNEDE VDAFLDLVEN ELTRLIEENS DLRQRINELD QELAAGGGAG VTPQAAQAIP AYEPEPGKPA PAAVSAGMNE EQALKAARVL SLAQDTADRL TNTAKAESDK MLADARANAE QILGEARHTA DATVAEARQR ...Sequence: MHHHHHHENL YFQGPLTPAD VHNVAFSKPP IGKRGYNEDE VDAFLDLVEN ELTRLIEENS DLRQRINELD QELAAGGGAG VTPQAAQAIP AYEPEPGKPA PAAVSAGMNE EQALKAARVL SLAQDTADRL TNTAKAESDK MLADARANAE QILGEARHTA DATVAEARQR ADAMLADAQS RSEAQLRQAQ EKADALQADA ERKHSEIMGT INQQRAVLEG RLEQLRTFER EYRTRLKTYL ESQLEELGQR GSAAPVDSNA DAGGFDQFNR GKN |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 2.87 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 2.87 mm | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||
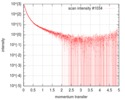
| Scan | Measurement date: Dec 15, 2017 / Storage temperature: 4 °C / Cell temperature: 4 °C / Exposure time: 1 sec. / Number of frames: 10 / Unit: 1/nm /
| ||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||
| Result | Comments: The cell wall synthesis protein Wag31: Polymer tbWag31 T73A forms a large super-macromolecular assembly (supported by atomic force microscopy, AFM) and as a result there is a very limited ...Comments: The cell wall synthesis protein Wag31: Polymer tbWag31 T73A forms a large super-macromolecular assembly (supported by atomic force microscopy, AFM) and as a result there is a very limited angular range to estimate the radius of gyration, Rg, using the Guinier approximation. In addition, the quoted monomer molecular weight does not apply to this sample. The probable distribution of real-space scattering pair vector lengths (p(r) vs r profile or PDDF) represents the cross-sectional pair distribution function for rod-like particle (option 4 in GNOM) and the subsequent data validation tools for this entry are based on the corresponding cross section parameters (Rg of cross section and Dmax of cross section). The I(s) is recorded in a relative scale in kDa units assuming a 1 mg/ml sample concentration. However the experimental concentration was found to be 2.7 mg/ml (assessed using a Bradford assay).
|
 Movie
Movie Controller
Controller