+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Protein translocase subunit SecA (full length, amino acids 1-901)
|
| Function / homology |  Function and homology information Function and homology informationpreprotein binding / cell envelope Sec protein transport complex / protein-exporting ATPase activity / protein-secreting ATPase / protein transport by the Sec complex / intracellular protein transmembrane transport / protein import / protein targeting to membrane / : / protein secretion ...preprotein binding / cell envelope Sec protein transport complex / protein-exporting ATPase activity / protein-secreting ATPase / protein transport by the Sec complex / intracellular protein transmembrane transport / protein import / protein targeting to membrane / : / protein secretion / ribonucleoprotein complex binding / protein targeting / cytoplasmic side of plasma membrane / protein transport / ribosome binding / zinc ion binding / ATP binding / identical protein binding / plasma membrane / cytoplasm Similarity search - Function |
| Biological species |  |
 Citation Citation |  Date: 2019 Jun 27 Date: 2019 Jun 27Title: The C-terminal tail of the bacterial translocation ATPase SecA modulates its activity Authors: Jamshad M / Knowles T / White S / Ward D / Mohammed F / Rahman K / Wynne M / Hughes G / Kramer G / Bukau B |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDDY9 SASDDY9 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
-Models
- Sample
Sample
 Sample Sample | Name: Protein translocase subunit SecA (full length, amino acids 1-901) Specimen concentration: 5 mg/ml |
|---|---|
| Buffer | Name: 20mM HEPES, 100mM NaCl, 1mM TCEP / pH: 8 |
| Entity #1174 | Type: protein / Description: Protein translocase subunit SecA / Formula weight: 102.022 / Num. of mol.: 2 / Source: Escherichia coli (strain K12) / References: UniProt: P10408 Sequence: MLIKLLTKVF GSRNDRTLRR MRKVVNIINA MEPEMEKLSD EELKGKTAEF RARLEKGEVL ENLIPEAFAV VREASKRVFG MRHFDVQLLG GMVLNERCIA EMRTGEGKTL TATLPAYLNA LTGKGVHVVT VNDYLAQRDA ENNRPLFEFL GLTVGINLPG MPAPAKREAY ...Sequence: MLIKLLTKVF GSRNDRTLRR MRKVVNIINA MEPEMEKLSD EELKGKTAEF RARLEKGEVL ENLIPEAFAV VREASKRVFG MRHFDVQLLG GMVLNERCIA EMRTGEGKTL TATLPAYLNA LTGKGVHVVT VNDYLAQRDA ENNRPLFEFL GLTVGINLPG MPAPAKREAY AADITYGTNN EYGFDYLRDN MAFSPEERVQ RKLHYALVDE VDSILIDEAR TPLIISGPAE DSSEMYKRVN KIIPHLIRQE KEDSETFQGE GHFSVDEKSR QVNLTERGLV LIEELLVKEG IMDEGESLYS PANIMLMHHV TAALRAHALF TRDVDYIVKD GEVIIVDEHT GRTMQGRRWS DGLHQAVEAK EGVQIQNENQ TLASITFQNY FRLYEKLAGM TGTADTEAFE FSSIYKLDTV VVPTNRPMIR KDLPDLVYMT EAEKIQAIIE DIKERTAKGQ PVLVGTISIE KSELVSNELT KAGIKHNVLN AKFHANEAAI VAQAGYPAAV TIATNMAGRG TDIVLGGSWQ AEVAALENPT AEQIEKIKAD WQVRHDAVLE AGGLHIIGTE RHESRRIDNQ LRGRSGRQGD AGSSRFYLSM EDALMRIFAS DRVSGMMRKL GMKPGEAIEH PWVTKAIANA QRKVESRNFD IRKQLLEYDD VANDQRRAIY SQRNELLDVS DVSETINSIR EDVFKATIDA YIPPQSLEEM WDIPGLQERL KNDFDLDLPI AEWLDKEPEL HEETLRERIL AQSIEVYQRK EEVVGAEMMR HFEKGVMLQT LDSLWKEHLA AMDYLRQGIH LRGYAQKDPK QEYKRESFSM FAAMLESLKY EVISTLSKVQ VRMPEEVEEL EQQRRMEAER LAQMQQLSHQ DDDSAAAAAL AAQTGERKVG RNDPCPCGSG KKYKQCHGRL Q |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.0919 Å / Dist. spec. to detc.: 2.867 mm / Type of source: X-ray synchrotron / Wavelength: 0.0919 Å / Dist. spec. to detc.: 2.867 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
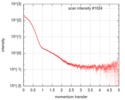
| Scan | Measurement date: Jul 18, 2016 / Storage temperature: 4 °C / Cell temperature: 20 °C / Exposure time: 1 sec. / Number of frames: 45 / Unit: 1/nm /
| |||||||||||||||||||||||||||||||||
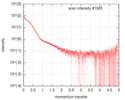
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result | Comments: Additional SEC parameters: Column type, S200 10/300 GL (GE Lifesciences); Flow rate, 1.0mL/min; Sample injection concentration, 5mg/mL; Injection volume, 0.5mL
|
 Movie
Movie Controller
Controller