[English] 日本語
 Yorodumi
Yorodumi- SASDDV7: p-hydroxyphenylacetate 3-hydroxylase, reductase component (mutant... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | p-hydroxyphenylacetate 3-hydroxylase, reductase component (mutant): 4 mg/ml of E248A C1 in the absence of p-hydroxyphenylacetic acid (HPA)
|
| Function / homology |  Function and homology information Function and homology informationflavin reductase (NADH) / flavin reductase (NADH) activity / riboflavin reductase (NADPH) activity / catabolic process / FMN binding Similarity search - Function |
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
 Citation Citation |  Journal: Arch Biochem Biophys / Year: 2018 Journal: Arch Biochem Biophys / Year: 2018Title: Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands. Authors: Anan Yuenyao / Nopphon Petchyam / Nuntaporn Kamonsutthipaijit / Pimchai Chaiyen / Danaya Pakotiprapha /  Abstract: The first step in the degradation of p-hydroxyphenylacetic acid (HPA) is catalyzed by the two-component enzyme p-hydroxyphenylacetate 3-hydroxylase (HPAH). The two components of Acinetobacter ...The first step in the degradation of p-hydroxyphenylacetic acid (HPA) is catalyzed by the two-component enzyme p-hydroxyphenylacetate 3-hydroxylase (HPAH). The two components of Acinetobacter baumannii HPAH are known as C and C, respectively. C is a flavin reductase that uses NADH to generate reduced flavin mononucleotide (FMNH), which is used by C in the hydroxylation of HPA. Interestingly, although HPA is not directly involved in the reaction catalyzed by C, the presence of HPA dramatically increases the FMN reduction rate. Amino acid sequence analysis revealed that C contains two domains: an N-terminal flavin reductase domain, and a C-terminal MarR domain. Although MarR proteins typically function as transcription regulators, the MarR domain of C was found to play an auto-inhibitory role. Here, we report a crystal structure of C and small-angle X-ray scattering (SAXS) studies that revealed that C undergoes a substantial conformational change in the presence of HPA, concomitant with the increase in the rate of flavin reduction. Amino acid residues that are important for HPA binding and regulation of C activity were identified by site-directed mutagenesis. Amino acid sequence similarity analysis revealed several as yet uncharacterized flavin reductases with N- or C-terminal fusions. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDDV7 SASDDV7 |
|---|
-Related structure data
| Related structure data |  5zc2C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
-Models
- Sample
Sample
 Sample Sample | Name: p-hydroxyphenylacetate 3-hydroxylase, reductase component (mutant): 4 mg/ml of E248A C1 in the absence of p-hydroxyphenylacetic acid (HPA) Specimen concentration: 4 mg/ml |
|---|---|
| Buffer | Name: 50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 10 % glycerol pH: 7 |
| Entity #1095 | Name: E248A C1 / Type: protein Description: p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E248A mutant Formula weight: 35.354 / Num. of mol.: 2 / Source: Acinetobacter baumannii / References: UniProt: Q6Q271 Sequence: MNQLNTAIVE KEVIDPMAFR RALGNFATGV TIMTAQTSSG ERVGVTANSF NSVSLDPALV LWSIDKKSSS YRIFEEATHF GVNILSAAQI ELSNRFARRS EDKFANIEFD LGVGNIPLFK NCSAAFECER YNIVEGGDHW IIIGRVVKFH DHGRSPLLYH QGAYSAVLPH ...Sequence: MNQLNTAIVE KEVIDPMAFR RALGNFATGV TIMTAQTSSG ERVGVTANSF NSVSLDPALV LWSIDKKSSS YRIFEEATHF GVNILSAAQI ELSNRFARRS EDKFANIEFD LGVGNIPLFK NCSAAFECER YNIVEGGDHW IIIGRVVKFH DHGRSPLLYH QGAYSAVLPH PSLNMKSETA EGVFPGRLYD NMYYLLTQAV RAYQNDYQPK QLASGFRTSE ARLLLVLESK TASSKCDLQR EVAMPIRAIE EATKILSEKG LLIDNGQHYE LTEQGNACAH MLYKIAESHQ EEVFAKYTVD ERKLFKNMLK DLIGI |
-Experimental information
| Beam | Instrument name: Synchrotron Light Research Institute (SLRI) BL1.3W City: Nakhon Ratchasima / 国: Thailand  / Type of source: X-ray synchrotron / Wavelength: 0.13776 Å / Dist. spec. to detc.: 1.332 mm / Type of source: X-ray synchrotron / Wavelength: 0.13776 Å / Dist. spec. to detc.: 1.332 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Rayonix SX165 / Type: CCD / Pixsize x: 15 mm | ||||||||||||||||||||||||||||||
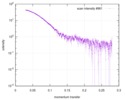
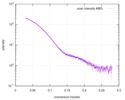
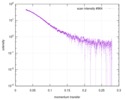
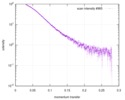
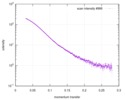
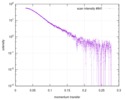
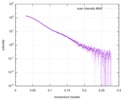
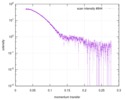
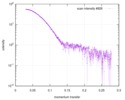
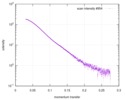
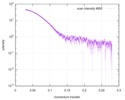
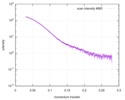
| Scan | Measurement date: Apr 25, 2018 / Storage temperature: 4 °C / Cell temperature: 16 °C / Exposure time: 600 sec. / Number of frames: 1 / Unit: 1/A /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller