+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: SASBDB / ID: SASDDM9 |
|---|---|
 試料 試料 | lipoprotein lipase–GPIHBP1 complex
|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報lipoprotein lipase / low-density lipoprotein particle mediated signaling / lipoprotein lipase activity / chylomicron remodeling / positive regulation of cholesterol storage / phospholipase A1 / glycerophospholipase activity / glycerophospholipid phospholipase A1 activity / Assembly of active LPL and LIPC lipase complexes / triglyceride catabolic process ...lipoprotein lipase / low-density lipoprotein particle mediated signaling / lipoprotein lipase activity / chylomicron remodeling / positive regulation of cholesterol storage / phospholipase A1 / glycerophospholipase activity / glycerophospholipid phospholipase A1 activity / Assembly of active LPL and LIPC lipase complexes / triglyceride catabolic process / very-low-density lipoprotein particle remodeling / Chylomicron remodeling / very-low-density lipoprotein particle clearance / chylomicron / positive regulation of lipid storage / high-density lipoprotein particle remodeling / cellular response to nutrient / triacylglycerol lipase activity / very-low-density lipoprotein particle / heparan sulfate proteoglycan binding / positive regulation of macrophage derived foam cell differentiation / cellular response to fatty acid / positive regulation of chemokine (C-X-C motif) ligand 2 production / triglyceride homeostasis / triglyceride metabolic process / lipoprotein particle binding / apolipoprotein binding / positive regulation of fat cell differentiation / response to glucose / catalytic complex / retinoid metabolic process / Retinoid metabolism and transport / positive regulation of chemokine production / protein-membrane adaptor activity / positive regulation of adipose tissue development / phospholipid metabolic process / cholesterol homeostasis / positive regulation of interleukin-1 beta production / response to bacterium / fatty acid metabolic process / Transcriptional regulation of white adipocyte differentiation / positive regulation of interleukin-6 production / positive regulation of tumor necrosis factor production / positive regulation of inflammatory response / fatty acid biosynthetic process / heparin binding / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / signaling receptor binding / calcium ion binding / cell surface / protein homodimerization activity / extracellular space / extracellular region / plasma membrane 類似検索 - 分子機能 |
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
 引用 引用 |  ジャーナル: Proc Natl Acad Sci U S A / 年: 2019 ジャーナル: Proc Natl Acad Sci U S A / 年: 2019タイトル: Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. 著者: Gabriel Birrane / Anne P Beigneux / Brian Dwyer / Bettina Strack-Logue / Kristian Kølby Kristensen / Omar L Francone / Loren G Fong / Haydyn D T Mertens / Clark Q Pan / Michael Ploug / ...著者: Gabriel Birrane / Anne P Beigneux / Brian Dwyer / Bettina Strack-Logue / Kristian Kølby Kristensen / Omar L Francone / Loren G Fong / Haydyn D T Mertens / Clark Q Pan / Michael Ploug / Stephen G Young / Muthuraman Meiyappan /    要旨: Lipoprotein lipase (LPL) is responsible for the intravascular processing of triglyceride-rich lipoproteins. The LPL within capillaries is bound to GPIHBP1, an endothelial cell protein with a three- ...Lipoprotein lipase (LPL) is responsible for the intravascular processing of triglyceride-rich lipoproteins. The LPL within capillaries is bound to GPIHBP1, an endothelial cell protein with a three-fingered LU domain and an N-terminal intrinsically disordered acidic domain. Loss-of-function mutations in or cause severe hypertriglyceridemia (chylomicronemia), but structures for LPL and GPIHBP1 have remained elusive. Inspired by our recent discovery that GPIHBP1's acidic domain preserves LPL structure and activity, we crystallized an LPL-GPIHBP1 complex and solved its structure. GPIHBP1's LU domain binds to LPL's C-terminal domain, largely by hydrophobic interactions. Analysis of electrostatic surfaces revealed that LPL contains a large basic patch spanning its N- and C-terminal domains. GPIHBP1's acidic domain was not defined in the electron density map but was positioned to interact with LPL's large basic patch, providing a likely explanation for how GPIHBP1 stabilizes LPL. The LPL-GPIHBP1 structure provides insights into mutations causing chylomicronemia. |
 登録者 登録者 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-モデル
| モデル #2159 | 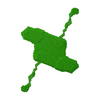 タイプ: dummy / ソフトウェア: (2.8.1) / ダミー原子の半径: 2.20 A / 対称性: P2 / カイ2乗値: 1.067 / P-value: 0.382446  Omokage検索でこの集合体の類似形状データを探す (詳細) Omokage検索でこの集合体の類似形状データを探す (詳細) |
|---|
- 試料
試料
 試料 試料 | 名称: lipoprotein lipase–GPIHBP1 complex / 試料濃度: 0.72 mg/ml / Entity id: 1184 / 1185 |
|---|---|
| バッファ | 名称: 10 mM BisTris, 150 mM NaCl, 4 mM CaCl2, 10% glycerol pH: 7.4 |
| 要素 #1184 | 名称: hLPL / タイプ: protein / 記述: Lipoprotein lipase / 分子量: 50.323 / 分子数: 2 / 由来: Homo sapiens / 参照: UniProt: P06858 配列: DQRRDFIDIE SKFALRTPED TAEDTCHLIP GVAESVATCH FNHSSKTFMV IHGWTVTGMY ESWVPKLVAA LYKREPDSNV IVVDWLSRAQ EHYPVSAGYT KLVGQDVARF INWMEEEFNY PLDNVHLLGY SLGAHAAGIA GSLTNKKVNR ITGLDPAGPN FEYAEAPSRL ...配列: DQRRDFIDIE SKFALRTPED TAEDTCHLIP GVAESVATCH FNHSSKTFMV IHGWTVTGMY ESWVPKLVAA LYKREPDSNV IVVDWLSRAQ EHYPVSAGYT KLVGQDVARF INWMEEEFNY PLDNVHLLGY SLGAHAAGIA GSLTNKKVNR ITGLDPAGPN FEYAEAPSRL SPDDADFVDV LHTFTRGSPG RSIGIQKPVG HVDIYPNGGT FQPGCNIGEA IRVIAERGLG DVDQLVKCSH ERSIHLFIDS LLNEENPSKA YRCSSKEAFE KGLCLSCRKN RCNNLGYEIN KVRAKRSSKM YLKTRSQMPY KVFHYQVKIH FSGTESETHT NQAFEISLYG TVAESENIPF TLPEVSTNKT YSFLIYTEVD IGELLMLKLK WKSDSYFSWS DWWSSPGFAI QKIRVKAGET QKKVIFCSRE KVSHLQKGKA PAVFVKCHDK SLNKKSG |
| 要素 #1185 | 名称: GPIHBP1 / タイプ: protein 記述: Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 分子量: 14.713 / 分子数: 2 / 由来: Homo sapiens 配列: QTQQEEEEED EDHGPDDYDE EDEDEVEEEE TNRLPGGRSR VLLRCYTCKS LPRDERCNLT QNCSHGQTCT TLIAHGNTES GLLTTHSTWC TDSCQPITKT VEGTQVTMTC CQSSLCNVPP WQSSRVQDPT G |
-実験情報
| ビーム | 設備名称: PETRA III EMBL P12 / 地域: Hamburg / 国: Germany  / 線源: X-ray synchrotron / 波長: 0.124 Å / スペクトロメータ・検出器間距離: 3.1 mm / 線源: X-ray synchrotron / 波長: 0.124 Å / スペクトロメータ・検出器間距離: 3.1 mm | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 検出器 | 名称: Pilatus 2M | ||||||||||||||||||||||||||||||||||||||||||
| スキャン |
| ||||||||||||||||||||||||||||||||||||||||||
| 距離分布関数 P(R) |
| ||||||||||||||||||||||||||||||||||||||||||
| 結果 |
|
 ムービー
ムービー コントローラー
コントローラー


 SASDDM9
SASDDM9




