[English] 日本語
 Yorodumi
Yorodumi- SASDDC6: Small glutamine-rich tetratricopeptide repeat-containing protein ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 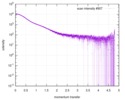 |
|---|---|
 Sample Sample | Small glutamine-rich tetratricopeptide repeat-containing protein alpha (N-terminal-TPR domains; SGTA_NT)
|
| Function / homology |  Function and homology information Function and homology informationTRC complex / negative regulation of ERAD pathway / tail-anchored membrane protein insertion into ER membrane / positive regulation of ERAD pathway / post-translational protein targeting to endoplasmic reticulum membrane / BAT3 complex binding / positive regulation of ubiquitin-dependent protein catabolic process / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / negative regulation of ubiquitin-dependent protein catabolic process / ERAD pathway ...TRC complex / negative regulation of ERAD pathway / tail-anchored membrane protein insertion into ER membrane / positive regulation of ERAD pathway / post-translational protein targeting to endoplasmic reticulum membrane / BAT3 complex binding / positive regulation of ubiquitin-dependent protein catabolic process / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / negative regulation of ubiquitin-dependent protein catabolic process / ERAD pathway / molecular adaptor activity / nucleoplasm / identical protein binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: BMC Biol / Year: 2018 Journal: BMC Biol / Year: 2018Title: Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control. Authors: Santiago Martínez-Lumbreras / Ewelina M Krysztofinska / Arjun Thapaliya / Alessandro Spilotros / Dijana Matak-Vinkovic / Enrico Salvadori / Peristera Roboti / Yvonne Nyathi / Janina H ...Authors: Santiago Martínez-Lumbreras / Ewelina M Krysztofinska / Arjun Thapaliya / Alessandro Spilotros / Dijana Matak-Vinkovic / Enrico Salvadori / Peristera Roboti / Yvonne Nyathi / Janina H Muench / Maxie M Roessler / Dmitri I Svergun / Stephen High / Rivka L Isaacson /   Abstract: BACKGROUND: Protein quality control mechanisms are essential for cell health and involve delivery of proteins to specific cellular compartments for recycling or degradation. In particular, stray ...BACKGROUND: Protein quality control mechanisms are essential for cell health and involve delivery of proteins to specific cellular compartments for recycling or degradation. In particular, stray hydrophobic proteins are captured in the aqueous cytosol by a co-chaperone, the small glutamine-rich, tetratricopeptide repeat-containing protein alpha (SGTA), which facilitates the correct targeting of tail-anchored membrane proteins, as well as the sorting of membrane and secretory proteins that mislocalize to the cytosol and endoplasmic reticulum-associated degradation. Full-length SGTA has an unusual elongated dimeric structure that has, until now, evaded detailed structural analysis. The C-terminal region of SGTA plays a key role in binding a broad range of hydrophobic substrates, yet in contrast to the well-characterized N-terminal and TPR domains, there is a lack of structural information on the C-terminal domain. In this study, we present new insights into the conformation and organization of distinct domains of SGTA and show that the C-terminal domain possesses a conserved region essential for substrate processing in vivo. RESULTS: We show that the C-terminal domain region is characterized by α-helical propensity and an intrinsic ability to dimerize independently of the N-terminal domain. Based on the properties of ...RESULTS: We show that the C-terminal domain region is characterized by α-helical propensity and an intrinsic ability to dimerize independently of the N-terminal domain. Based on the properties of different regions of SGTA that are revealed using cell biology, NMR, SAXS, Native MS, and EPR, we observe that its C-terminal domain can dimerize in the full-length protein and propose that this reflects a closed conformation of the substrate-binding domain. CONCLUSION: Our results provide novel insights into the structural complexity of SGTA and provide a new basis for mechanistic studies of substrate binding and release at the C-terminal region. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDDC6 SASDDC6 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Small glutamine-rich tetratricopeptide repeat-containing protein alpha (N-terminal-TPR domains; SGTA_NT) Specimen concentration: 0.60-4.50 |
|---|---|
| Buffer | Name: 10 mM potassium phosphate, 100 mM NaCl / pH: 6 |
| Entity #1056 | Name: SGTA_NT-TPR / Type: protein Description: Small glutamine-rich tetratricopeptide repeat-containing protein alpha Nterminal-TPR domains Formula weight: 23.592 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: O43765 Sequence: GSMDNKKRLA YAIIQFLHDQ LRHGGLSSDA QESLEVAIQC LETAFGVTVE DSDLALPQTL PEIFEAAATG KEMPQDLRSP ARTPPSEEDS AEAERLKTEG NEQMKVENFE AAVHFYGKAI ELNPANAVYF CNRAAAYSKL GNYAGAVQDC ERAICIDPAY SKAYGRMGLA ...Sequence: GSMDNKKRLA YAIIQFLHDQ LRHGGLSSDA QESLEVAIQC LETAFGVTVE DSDLALPQTL PEIFEAAATG KEMPQDLRSP ARTPPSEEDS AEAERLKTEG NEQMKVENFE AAVHFYGKAI ELNPANAVYF CNRAAAYSKL GNYAGAVQDC ERAICIDPAY SKAYGRMGLA LSSLNKHVEA VAYYKKALEL DPDNETYKSN LKIAELKLRE APSPT |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||
| Scan | Measurement date: Jun 5, 2015 / Cell temperature: 25 °C / Exposure time: 0.045 sec. / Number of frames: 20 / Unit: 1/nm /
| |||||||||||||||
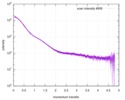
| Result | Experimental MW: 41 kDa / Type of curve: merged
|
 Movie
Movie Controller
Controller