[English] 日本語
 Yorodumi
Yorodumi- SASDB24: Aureochrome 1a bZIP-LOV module: PtAUREO1a bZIP-LOV (Light oxygen ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 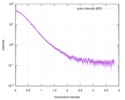 |
|---|---|
 Sample Sample | Aureochrome 1a bZIP-LOV module: PtAUREO1a bZIP-LOV (Light oxygen voltage) module (light state, Tris)
|
| Biological species |  |
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2016 Journal: Nucleic Acids Res / Year: 2016Title: Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes. Authors: Ankan Banerjee / Elena Herman / Manuel Serif / Manuel Maestre-Reyna / Sebastian Hepp / Richard Pokorny / Peter G Kroth / Lars-Oliver Essen / Tilman Kottke /   Abstract: The modular architecture of aureochrome blue light receptors, found in several algal groups including diatoms, is unique by having the LOV-type photoreceptor domain fused to the C-terminus of its ...The modular architecture of aureochrome blue light receptors, found in several algal groups including diatoms, is unique by having the LOV-type photoreceptor domain fused to the C-terminus of its putative effector, an N-terminal DNA-binding bZIP module. The structural and functional understanding of aureochromes' light-dependent signaling mechanism is limited, despite their promise as an optogenetic tool. We show that class I aureochromes 1a and 1c from the diatom Phaeodactylum tricornutum are regulated in a light-independent circadian rhythm. These aureochromes are capable to form functional homo- and heterodimers, which recognize the ACGT core sequence within the canonical 'aureo box', TGACGT, in a light-independent manner. The bZIP domain holds a more folded and less flexible but extended conformation in the duplex DNA-bound state. FT-IR spectroscopy in the absence and the presence of DNA shows light-dependent helix unfolding in the LOV domain, which leads to conformational changes in the bZIP region. The solution structure of DNA bound to aureochrome points to a tilted orientation that was further validated by molecular dynamics simulations. We propose that aureochrome signaling relies on an allosteric pathway from LOV to bZIP that results in conformational changes near the bZIP-DNA interface without major effects on the binding affinity. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Aureochrome 1a bZIP-LOV module: PtAUREO1a bZIP-LOV (Light oxygen voltage) module (light state, Tris) Specimen concentration: 0.63-2.50 |
|---|---|
| Buffer | Name: Tris / Concentration: 10.00 mM / pH: 8 / Composition: 300 mM NaCl |
| Entity #312 | Name: PtAUREO1a bZIP-LOV / Type: protein / Description: Aureochrome 1a bZIP-LOV module / Formula weight: 28.655 / Num. of mol.: 2 / Source: Phaeodactylum tricornutum Sequence: MGSSHHHHHH SSGLVPRGSH MDRKMSEQQK VERRERNREH AKRSRIRKKF LLESLQQSVS LLKEENEKLK TSIRSHLGEE KADTLIDSAN NNKTDVDGLL ASSQGIANKV LDDPDFSFIK ALQTAQQNFV VTDPSLPDNP IVYASQGFLN LTGYSLDQIL GRNCRFLQGP ...Sequence: MGSSHHHHHH SSGLVPRGSH MDRKMSEQQK VERRERNREH AKRSRIRKKF LLESLQQSVS LLKEENEKLK TSIRSHLGEE KADTLIDSAN NNKTDVDGLL ASSQGIANKV LDDPDFSFIK ALQTAQQNFV VTDPSLPDNP IVYASQGFLN LTGYSLDQIL GRNCRFLQGP ETDPKAVERI RKAIEQGNDM SVCLLNYRVD GTTFWNQFFI AALRDAGGNV TNFVGVQCKV SDQYAATVTK QQEEEEEAAA NDDED |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.93 Å / Dist. spec. to detc.: 2.43 mm / Type of source: X-ray synchrotron / Wavelength: 0.93 Å / Dist. spec. to detc.: 2.43 mm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M | |||||||||||||||||||||||||||
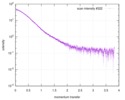
| Scan |
| |||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller About Yorodumi
About Yorodumi


 SASDB24
SASDB24