[English] 日本語
 Yorodumi
Yorodumi- SASDAZ7: IL-6R AIR-3A 2:4 complex (AIR-3A + Interleukin-6 receptor subunit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAZ7 |
|---|---|
 Sample Sample | IL-6R AIR-3A 2:4 complex
|
| Function / homology |  Function and homology information Function and homology informationciliary neurotrophic factor binding / hepatic immune response / interleukin-6 receptor activity / interleukin-6 binding / ciliary neurotrophic factor-mediated signaling pathway / interleukin-11 receptor activity / interleukin-11 binding / ciliary neurotrophic factor receptor complex / interleukin-6 receptor complex / T-helper 17 cell lineage commitment ...ciliary neurotrophic factor binding / hepatic immune response / interleukin-6 receptor activity / interleukin-6 binding / ciliary neurotrophic factor-mediated signaling pathway / interleukin-11 receptor activity / interleukin-11 binding / ciliary neurotrophic factor receptor complex / interleukin-6 receptor complex / T-helper 17 cell lineage commitment / endocrine pancreas development / negative regulation of collagen biosynthetic process / positive regulation of leukocyte chemotaxis / vascular endothelial growth factor production / positive regulation of glomerular mesangial cell proliferation / negative regulation of interleukin-8 production / neutrophil mediated immunity / cytokine receptor activity / cell surface receptor signaling pathway via STAT / Interleukin-6 signaling / interleukin-6-mediated signaling pathway / MAPK3 (ERK1) activation / MAPK1 (ERK2) activation / monocyte chemotaxis / positive regulation of osteoblast differentiation / positive regulation of chemokine production / extrinsic apoptotic signaling pathway / response to cytokine / positive regulation of smooth muscle cell proliferation / acute-phase response / positive regulation of interleukin-6 production / Transcriptional regulation of granulopoiesis / cytokine-mediated signaling pathway / Interleukin-4 and Interleukin-13 signaling / Potential therapeutics for SARS / defense response to Gram-negative bacterium / receptor complex / positive regulation of MAPK cascade / apical plasma membrane / defense response to Gram-positive bacterium / external side of plasma membrane / positive regulation of cell population proliferation / enzyme binding / protein homodimerization activity / extracellular space / extracellular region / plasma membrane Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: RNA Biol / Year: 2016 Journal: RNA Biol / Year: 2016Title: Structure and target interaction of a G-quadruplex RNA-aptamer. Authors: Kristina Szameit / Katharina Berg / Sven Kruspe / Erica Valentini / Eileen Magbanua / Marcel Kwiatkowski / Isaure Chauvot de Beauchêne / Boris Krichel / Kira Schamoni / Charlotte Uetrecht / ...Authors: Kristina Szameit / Katharina Berg / Sven Kruspe / Erica Valentini / Eileen Magbanua / Marcel Kwiatkowski / Isaure Chauvot de Beauchêne / Boris Krichel / Kira Schamoni / Charlotte Uetrecht / Dmitri I Svergun / Hartmut Schlüter / Martin Zacharias / Ulrich Hahn /  Abstract: G-quadruplexes have recently moved into focus of research in nucleic acids, thereby evolving in scientific significance from exceptional secondary structure motifs to complex modulators of gene ...G-quadruplexes have recently moved into focus of research in nucleic acids, thereby evolving in scientific significance from exceptional secondary structure motifs to complex modulators of gene regulation. Aptamers (nucleic acid based ligands with recognition properties for a specific target) that form Gquadruplexes may have particular potential for therapeutic applications as they combine the characteristics of specific targeting and Gquadruplex mediated stability and regulation. We have investigated the structure and target interaction properties of one such aptamer: AIR-3 and its truncated form AIR-3A. These RNA aptamers are specific for human interleukin-6 receptor (hIL-6R), a key player in inflammatory diseases and cancer, and have recently been exploited for in vitro drug delivery studies. With the aim to resolve the RNA structure, global shape, RNA:protein interaction site and binding stoichiometry, we now investigated AIR-3 and AIR-3A by different methods including RNA structure probing, Small Angle X-ray scattering and microscale thermophoresis. Our findings suggest a broader spectrum of folding species than assumed so far and remarkable tolerance toward different modifications. Mass spectrometry based binding site analysis, supported by molecular modeling and docking studies propose a general Gquadruplex affinity for the target molecule hIL-6R. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDAZ7 SASDAZ7 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
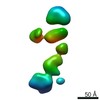
| Model #305 |  Type: dummy / Radius of dummy atoms: 5.50 A / Chi-square value: 1.036  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #306 |  Type: mix / Symmetry: P1 / Chi-square value: 1.43  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: IL-6R AIR-3A 2:4 complex / Specimen concentration: 0.2 mg/ml / Entity id: 173 / 174 |
|---|---|
| Buffer | Name: water / pH: 7.5 |
| Entity #173 | Type: RNA / Description: AIR-3A / Formula weight: 6.388 / Num. of mol.: 4 Sequence: GGGGAGGCUG UGGUGAGGG |
| Entity #174 | Name: IL-6R / Type: protein / Description: Interleukin-6 receptor subunit alpha / Formula weight: 40.964 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: P08887 Sequence: MRAWIFFLLC LAGRALAAPL VHHHHHHALA PRRCPAQEVA RGVLTSLPGD SVTLTCPGVE PEDNATVHWV LRKPAAGSHP SRWAGMGRRL LLRSVQLHDS GNYSCYRAGR PAGTVHLLVD VPPEEPQLSC FRKSPLSNVV CEWGPRSTPS LTTKAVLLVR KFQNSPAEDF ...Sequence: MRAWIFFLLC LAGRALAAPL VHHHHHHALA PRRCPAQEVA RGVLTSLPGD SVTLTCPGVE PEDNATVHWV LRKPAAGSHP SRWAGMGRRL LLRSVQLHDS GNYSCYRAGR PAGTVHLLVD VPPEEPQLSC FRKSPLSNVV CEWGPRSTPS LTTKAVLLVR KFQNSPAEDF QEPCQYSQES QKFSCQLAVP EGDSSFYIVS MCVASSVGSK FSKTQTFQGC GILQPDPPAN ITVTAVARNP RWLSVTWQDP HSWNSSFYRL RFELRYRAER SKTFTTWMVK DLQHHCVIHD AWSGLRHVVQ LRAQEEFGQG EWSEWSPEAM GTPWTESRSP PAENEVSTPM QALTTNKDDD NILFRDSANA TSLPVQ |
-Experimental information
| Beam | Instrument name: Diamond Light Source B21 / City: Oxfordshire / 国: UK  / Shape: 1 x 5 mm / Type of source: X-ray synchrotron / Dist. spec. to detc.: 3.9 mm / Shape: 1 x 5 mm / Type of source: X-ray synchrotron / Dist. spec. to detc.: 3.9 mm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||||||||
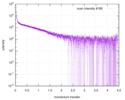
| Scan |
| |||||||||||||||||||||||||||
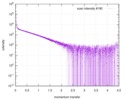
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||
| Result | Comments: The CORAL model fits the experimental curve with a Chi-squared-value of 1.43. The model appears slightly smaller probably because of the presence of free RNA dimers in solution.
|
 Movie
Movie Controller
Controller