+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8zmz | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of R-eLACCO2 in the lactate-bound state | ||||||||||||||||||||||||
 Components Components | Lactate-binding periplasmic protein TTHA0766,Red fluorescent protein drFP583 | ||||||||||||||||||||||||
 Keywords Keywords | FLUORESCENT PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationlactate transport / tripartite ATP-independent periplasmic transporter complex / bioluminescence / generation of precursor metabolites and energy / transmembrane transport / periplasmic space / calcium ion binding / protein homodimerization activity / zinc ion binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) Discosoma sp. (sea anemone) Discosoma sp. (sea anemone) | ||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||||||||||||||
 Authors Authors | Kamijo, Y. / Kusakizako, T. / Nureki, O. / Campbell, R.E. / Nasu, Y. | ||||||||||||||||||||||||
| Funding support |  Japan, 2items Japan, 2items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: A red fluorescent genetically encoded biosensor for in vivo imaging of extracellular L-lactate dynamics. Authors: Yuki Kamijo / Philipp Mächler / Natalie Ness / Cong Quang Vu / Tsukasa Kusakizako / Jamsad Mannuthodikayil / Zaneta Ku / Marc Boisvert / Ekaterina Grebenik / Ikumi Miyazaki / Rina Hashizume ...Authors: Yuki Kamijo / Philipp Mächler / Natalie Ness / Cong Quang Vu / Tsukasa Kusakizako / Jamsad Mannuthodikayil / Zaneta Ku / Marc Boisvert / Ekaterina Grebenik / Ikumi Miyazaki / Rina Hashizume / Haruaki Sato / Rui Liu / Yukiko Hori / Taisuke Tomita / Tetsuro Katayama / Akihiro Furube / Gabriela Caraveo / Marie-Eve Paquet / Mikhail Drobizhev / Osamu Nureki / Satoshi Arai / Marco Brancaccio / Robert E Campbell / David Kleinfeld / Yusuke Nasu /      Abstract: L-Lactate is increasingly recognized as an intercellular energy currency in mammals, but mysteries remain regarding the spatial and temporal dynamics of its release and uptake between cells via the ...L-Lactate is increasingly recognized as an intercellular energy currency in mammals, but mysteries remain regarding the spatial and temporal dynamics of its release and uptake between cells via the extracellular environment. Here we introduce R-eLACCO2.1, a red fluorescent extracellular L-lactate biosensor that is superior to previously reported green fluorescent biosensors in in vivo sensitivity to increases in extracellular L-lactate and spectral orthogonality. R-eLACCO2.1 exhibits excellent fluorescence response in cultured cells, mouse brain slices, and live mice. R-eLACCO2.1 also serves as an effective fluorescence lifetime-based biosensor. Using R-eLACCO2.1, we monitor whisker stimulation and locomotion-induced changes in endogenous extracellular L-lactate in the somatosensory cortex of awake mice. To highlight the potential insights gained from in vivo measurements with R-eLACCO2.1, we perform dual-color imaging from the somatosensory cortex of actively locomoting mice. This enables us to simultaneously observe the neural activity, reported by a green fluorescent GCaMP calcium ion biosensor, and extracellular L-lactate. As the high-performance tool in the suite of extracellular L-lactate biosensors, R-eLACCO2.1 is ideally suited to delimit the emerging roles of L-lactate in mammalian metabolism. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8zmz.cif.gz 8zmz.cif.gz | 156.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8zmz.ent.gz pdb8zmz.ent.gz | 94.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8zmz.json.gz 8zmz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8zmz_validation.pdf.gz 8zmz_validation.pdf.gz | 1.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8zmz_full_validation.pdf.gz 8zmz_full_validation.pdf.gz | 1.5 MB | Display | |
| Data in XML |  8zmz_validation.xml.gz 8zmz_validation.xml.gz | 36.1 KB | Display | |
| Data in CIF |  8zmz_validation.cif.gz 8zmz_validation.cif.gz | 51.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zm/8zmz https://data.pdbj.org/pub/pdb/validation_reports/zm/8zmz ftp://data.pdbj.org/pub/pdb/validation_reports/zm/8zmz ftp://data.pdbj.org/pub/pdb/validation_reports/zm/8zmz | HTTPS FTP |
-Related structure data
| Related structure data |  60263MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
| Symmetry | Point symmetry: (Schoenflies symbol: C2 (2 fold cyclic)) |
- Components
Components
| #1: Protein | Mass: 70044.500 Da / Num. of mol.: 1 / Mutation: mutant type Source method: isolated from a genetically manipulated source Details: Chimeric protein of THA0766 and cpmApple Source: (gene. exp.)   Thermus thermophilus HB8 (bacteria), (gene. exp.) Thermus thermophilus HB8 (bacteria), (gene. exp.)  Discosoma sp. (sea anemone) Discosoma sp. (sea anemone)Gene: TTHA0766 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-CA / |
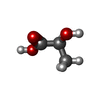
| #3: Chemical | ChemComp-2OP / ( |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: R-eLACCO2 in the lactate-bound state / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||
| Buffer solution | pH: 8 | ||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1600 nm / Nominal defocus min: 600 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||
| 3D reconstruction | Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 146454 / Symmetry type: POINT | |||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj