+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8u53 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Mechanically activated ion channel OSCA3.1 in nanodiscs | ||||||
 Components Components | CSC1-like protein ERD4 | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / Mechanically activated ion channel | ||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast envelope / plasmodesma / plant-type vacuole / mechanosensitive monoatomic ion channel activity / calcium-activated cation channel activity / mRNA binding / nucleus / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.6 Å | ||||||
 Authors Authors | Jojoa-Cruz, S. / Lee, W.H. / Ward, A.B. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli. Authors: Sebastian Jojoa-Cruz / Adrienne E Dubin / Wen-Hsin Lee / Andrew B Ward /  Abstract: The dimeric two-pore OSCA/TMEM63 family has recently been identified as mechanically activated ion channels. Previously, based on the unique features of the structure of OSCA1.2, we postulated the ...The dimeric two-pore OSCA/TMEM63 family has recently been identified as mechanically activated ion channels. Previously, based on the unique features of the structure of OSCA1.2, we postulated the potential involvement of several structural elements in sensing membrane tension (Jojoa-Cruz et al., 2018). Interestingly, while OSCA1, 2, and 3 clades are activated by membrane stretch in cell-attached patches (i.e. they are stretch-activated channels), they differ in their ability to transduce membrane deformation induced by a blunt probe (poking). Here, in an effort to understand the domains contributing to mechanical signal transduction, we used cryo-electron microscopy to solve the structure of (At) OSCA3.1, which, unlike AtOSCA1.2, only produced stretch- but not poke-activated currents in our initial characterization (Murthy et al., 2018). Mutagenesis and electrophysiological assessment of conserved and divergent putative mechanosensitive features of OSCA1.2 reveal a selective disruption of the macroscopic currents elicited by poking without considerable effects on stretch-activated currents (SAC). Our results support the involvement of the amphipathic helix and lipid-interacting residues in the membrane fenestration in the response to poking. Our findings position these two structural elements as potential sources of functional diversity within the family. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8u53.cif.gz 8u53.cif.gz | 246.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8u53.ent.gz pdb8u53.ent.gz | 197 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8u53.json.gz 8u53.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/u5/8u53 https://data.pdbj.org/pub/pdb/validation_reports/u5/8u53 ftp://data.pdbj.org/pub/pdb/validation_reports/u5/8u53 ftp://data.pdbj.org/pub/pdb/validation_reports/u5/8u53 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  41911MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
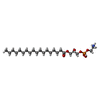
| #1: Protein | Mass: 81878.898 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: The last 10 residues (GTGTLEVLFQ) are leftover of a linker and protease site. Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: Q9C8G5 Homo sapiens (human) / References: UniProt: Q9C8G5#2: Chemical | #3: Chemical | #4: Chemical | ChemComp-PLM / Has ligand of interest | N | Sequence details | Residues that could not be identified in the map are represented as UNK. The actual sequence of the ...Residues that could not be identified in the map are represented as UNK. The actual sequence of the protein construct is: MEFGSFLVSL | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: OSCA3.1 dimer in nanodisc / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.164 MDa / Experimental value: YES | ||||||||||||||||||||
| Source (natural) | Organism:  | ||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) / Cell: HEK293F / Plasmid: pcDNA3.1 Homo sapiens (human) / Cell: HEK293F / Plasmid: pcDNA3.1 | ||||||||||||||||||||
| Buffer solution | pH: 8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 3.2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1500 nm / Nominal defocus min: 400 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 8375 Details: Only micrographs with a CTF estimate of 2.6A or better were used for processing |
| Image scans | Movie frames/image: 38 / Used frames/image: 1-38 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1913316 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.6 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 197944 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Accession code: 6MGV / Source name: SwissModel / Type: in silico model |
 Movie
Movie Controller
Controller



 PDBj
PDBj