[English] 日本語
 Yorodumi
Yorodumi- PDB-8rq4: Cryo-em structure of the rat Multidrug resistance-associated prot... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8rq4 | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-em structure of the rat Multidrug resistance-associated protein 2 (rMrp2) in complex with probenecid | |||||||||||||||||||||||||||||||||||||||||||||
 Components Components | ATP-binding cassette sub-family C member 2 | |||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / Multidrug resistance-associated protein 2 (rMrp2) in an autoinhibited state (nucleotide-free) | |||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmercury ion transport / benzylpenicillin metabolic process / Aspirin ADME / Atorvastatin ADME / Paracetamol ADME / antibiotic metabolic process / canalicular bile acid transport / intracellular canaliculus / bilirubin transmembrane transporter activity / bilirubin transport ...mercury ion transport / benzylpenicillin metabolic process / Aspirin ADME / Atorvastatin ADME / Paracetamol ADME / antibiotic metabolic process / canalicular bile acid transport / intracellular canaliculus / bilirubin transmembrane transporter activity / bilirubin transport / Heme degradation / xenobiotic export from cell / ABC-family proteins mediated transport / detoxification of mercury ion / intracellular chloride ion homeostasis / response to antineoplastic agent / thyroid hormone transport / leukotriene transport / prostaglandin transport / ABC-type glutathione S-conjugate transporter activity / ABC-type glutathione-S-conjugate transporter / xenobiotic transport across blood-brain barrier / xenobiotic transmembrane transport / organic anion transport / intercellular canaliculus / transepithelial transport / : / response to arsenic-containing substance / xenobiotic detoxification by transmembrane export across the plasma membrane / cellular response to interleukin-6 / ABC-type xenobiotic transporter / response to steroid hormone / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / response to glucagon / ABC-type xenobiotic transporter activity / bile acid and bile salt transport / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / cellular response to interleukin-1 / cellular response to dexamethasone stimulus / xenobiotic catabolic process / brush border membrane / female pregnancy / transmembrane transport / response to estrogen / cellular response to xenobiotic stimulus / cellular response to tumor necrosis factor / response to estradiol / cellular response to lipopolysaccharide / response to oxidative stress / response to lipopolysaccharide / apical plasma membrane / response to xenobiotic stimulus / protein domain specific binding / negative regulation of gene expression / cell surface / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Mazza, T. / Beis, K. | |||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Canada, 1items Canada, 1items
| |||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for the modulation of MRP2 activity by phosphorylation and drugs. Authors: Tiziano Mazza / Theodoros I Roumeliotis / Elena Garitta / David Drew / S Tamir Rashid / Cesare Indiveri / Jyoti S Choudhary / Kenneth J Linton / Konstantinos Beis /    Abstract: Multidrug resistance-associated protein 2 (MRP2/ABCC2) is a polyspecific efflux transporter of organic anions expressed in hepatocyte canalicular membranes. MRP2 dysfunction, in Dubin-Johnson ...Multidrug resistance-associated protein 2 (MRP2/ABCC2) is a polyspecific efflux transporter of organic anions expressed in hepatocyte canalicular membranes. MRP2 dysfunction, in Dubin-Johnson syndrome or by off-target inhibition, for example by the uricosuric drug probenecid, elevates circulating bilirubin glucuronide and is a cause of jaundice. Here, we determine the cryo-EM structure of rat Mrp2 (rMrp2) in an autoinhibited state and in complex with probenecid. The autoinhibited state exhibits an unusual conformation for this class of transporter in which the regulatory domain is folded within the transmembrane domain cavity. In vitro phosphorylation, mass spectrometry and transport assays show that phosphorylation of the regulatory domain relieves this autoinhibition and enhances rMrp2 transport activity. The in vitro data is confirmed in human hepatocyte-like cells, in which inhibition of endogenous kinases also reduces human MRP2 transport activity. The drug-bound state reveals two probenecid binding sites that suggest a dynamic interplay with autoinhibition. Mapping of the Dubin-Johnson mutations onto the rodent structure indicates that many may interfere with the transition between conformational states. | |||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8rq4.cif.gz 8rq4.cif.gz | 260.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8rq4.ent.gz pdb8rq4.ent.gz | 204.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8rq4.json.gz 8rq4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rq/8rq4 https://data.pdbj.org/pub/pdb/validation_reports/rq/8rq4 ftp://data.pdbj.org/pub/pdb/validation_reports/rq/8rq4 ftp://data.pdbj.org/pub/pdb/validation_reports/rq/8rq4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  19433MC  8rq3C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 173571.578 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q63120, Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate, ABC-type xenobiotic transporter, ABC-type ...References: UniProt: Q63120, Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate, ABC-type xenobiotic transporter, ABC-type glutathione-S-conjugate transporter | ||||
|---|---|---|---|---|---|
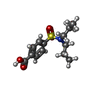
| #2: Chemical | ChemComp-Y01 / | ||||
| #3: Chemical | | Has ligand of interest | Y | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-em structure of the rat Multidrug resistance-associated protein 2 (rMrp2) in complex with probenecid Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2400 nm / Nominal defocus min: 1200 nm |
| Image recording | Electron dose: 52.8 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.45 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 247763 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL | ||||||||||||||||||||||||
| Atomic model building | PDB-ID: 8RQ3 Pdb chain-ID: A / Accession code: 8RQ3 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj