[English] 日本語
 Yorodumi
Yorodumi- PDB-8qo4: Conserved Structures and Dynamics in 5-Proximal Regions of Betaco... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8qo4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Components Components | MERS-CoV-SL5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | VIRUS / RNA structure / 5-proximal region / Coronavirus / Cryo-EM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology | : / RNA / RNA (> 10) / RNA (> 100) Function and homology information Function and homology information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 5.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Moura, T.R. / Purta, E. / Bernat, A. / Baulin, E. / Mukherjee, S. / Bujnicki, J.M. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Poland, 1items Poland, 1items
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes. Authors: Tales Rocha de Moura / Elżbieta Purta / Agata Bernat / Eva M Martín-Cuevas / Małgorzata Kurkowska / Eugene F Baulin / Sunandan Mukherjee / Jakub Nowak / Artur P Biela / Michał Rawski / ...Authors: Tales Rocha de Moura / Elżbieta Purta / Agata Bernat / Eva M Martín-Cuevas / Małgorzata Kurkowska / Eugene F Baulin / Sunandan Mukherjee / Jakub Nowak / Artur P Biela / Michał Rawski / Sebastian Glatt / Fernando Moreno-Herrero / Janusz M Bujnicki /   Abstract: Betacoronaviruses are a genus within the Coronaviridae family of RNA viruses. They are capable of infecting vertebrates and causing epidemics as well as global pandemics in humans. Mitigating the ...Betacoronaviruses are a genus within the Coronaviridae family of RNA viruses. They are capable of infecting vertebrates and causing epidemics as well as global pandemics in humans. Mitigating the threat posed by Betacoronaviruses requires an understanding of their molecular diversity. The development of novel antivirals hinges on understanding the key regulatory elements within the viral RNA genomes, in particular the 5'-proximal region, which is pivotal for viral protein synthesis. Using a combination of cryo-electron microscopy, atomic force microscopy, chemical probing, and computational modeling, we determined the structures of 5'-proximal regions in RNA genomes of Betacoronaviruses from four subgenera: OC43-CoV, SARS-CoV-2, MERS-CoV, and Rousettus bat-CoV. We obtained cryo-electron microscopy maps and determined atomic-resolution models for the stem-loop-5 (SL5) region at the translation start site and found that despite low sequence similarity and variable length of the helical elements it exhibits a remarkable structural conservation. Atomic force microscopy imaging revealed a common domain organization and a dynamic arrangement of structural elements connected with flexible linkers across all four Betacoronavirus subgenera. Together, these results reveal common features of a critical regulatory region shared between different Betacoronavirus RNA genomes, which may allow targeting of these RNAs by broad-spectrum antiviral therapeutics. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8qo4.cif.gz 8qo4.cif.gz | 628.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8qo4.ent.gz pdb8qo4.ent.gz | 539.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8qo4.json.gz 8qo4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qo/8qo4 https://data.pdbj.org/pub/pdb/validation_reports/qo/8qo4 ftp://data.pdbj.org/pub/pdb/validation_reports/qo/8qo4 ftp://data.pdbj.org/pub/pdb/validation_reports/qo/8qo4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  18522MC  8qo2C 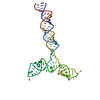 8qo3C  8qo5C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: RNA chain | Mass: 50119.512 Da / Num. of mol.: 1 / Source method: obtained synthetically Source: (synth.)  References: GenBank: NC_019843.3 |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Middle East respiratory syndrome-related coronavirus / Type: VIRUS / Details: in vitro transcription / Entity ID: all / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  | ||||||||||||||||||||
| Source (recombinant) | Organism: synthetic construct (others) | ||||||||||||||||||||
| Details of virus | Empty: YES / Enveloped: YES / Isolate: OTHER / Type: VIROID | ||||||||||||||||||||
| Natural host | Organism: synthetic construct | ||||||||||||||||||||
| Buffer solution | pH: 7.5 / Details: 20 mM HEPES pH 7.5, 50 mM KCl, 50 mM NaCl | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 0.9 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK II / Cryogen name: ETHANE / Humidity: 90 % / Chamber temperature: 298 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Nominal defocus max: 2100 nm / Nominal defocus min: 900 nm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 40 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 5.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 29234 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj





























