[English] 日本語
 Yorodumi
Yorodumi- PDB-8hbh: Structure of human soluble guanylate cyclase in the NO-activated ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8hbh | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human soluble guanylate cyclase in the NO-activated state at 3.1 angstrom | |||||||||||||||||||||
 Components Components | (Guanylate cyclase soluble subunit ...) x 2 | |||||||||||||||||||||
 Keywords Keywords | SIGNALING PROTEIN / soluble guanylate cyclase | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationretrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission / cytidylate cyclase activity / guanylate cyclase complex, soluble / guanylate cyclase / cGMP biosynthetic process / guanylate cyclase activity / presynaptic active zone cytoplasmic component / response to oxygen levels / nitric oxide binding / relaxation of vascular associated smooth muscle ...retrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission / cytidylate cyclase activity / guanylate cyclase complex, soluble / guanylate cyclase / cGMP biosynthetic process / guanylate cyclase activity / presynaptic active zone cytoplasmic component / response to oxygen levels / nitric oxide binding / relaxation of vascular associated smooth muscle / Nitric oxide stimulates guanylate cyclase / adenylate cyclase activity / blood circulation / : / positive regulation of nitric oxide mediated signal transduction / nitric oxide mediated signal transduction / Smooth Muscle Contraction / nitric oxide-cGMP-mediated signaling / cellular response to nitric oxide / Hsp90 protein binding / GABA-ergic synapse / regulation of blood pressure / signaling receptor activity / heme binding / GTP binding / protein-containing complex binding / glutamatergic synapse / metal ion binding / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||||||||
 Authors Authors | Chen, L. / Liu, R. | |||||||||||||||||||||
| Funding support |  China, 3items China, 3items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nitric Oxide / Year: 2023 Journal: Nitric Oxide / Year: 2023Title: NO binds to the distal site of haem in the fully activated soluble guanylate cyclase. Authors: Rui Liu / Yunlu Kang / Lei Chen /  Abstract: Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO). The binding of NO to the haem of sGC induces a large conformational change in the enzyme and activates its cyclase ...Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO). The binding of NO to the haem of sGC induces a large conformational change in the enzyme and activates its cyclase activity. However, whether NO binds to the proximal site or the distal site of haem in the fully activated state remains under debate. Here, we present cryo-EM maps of sGC in the NO-activated state at high resolutions, allowing the observation of the density of NO. These cryo-EM maps show the binding of NO to the distal site of haem in the NO-activated state. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8hbh.cif.gz 8hbh.cif.gz | 231.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8hbh.ent.gz pdb8hbh.ent.gz | 176.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8hbh.json.gz 8hbh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hb/8hbh https://data.pdbj.org/pub/pdb/validation_reports/hb/8hbh ftp://data.pdbj.org/pub/pdb/validation_reports/hb/8hbh ftp://data.pdbj.org/pub/pdb/validation_reports/hb/8hbh | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  34632MC  8hbeC  8hbfC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanylate cyclase soluble subunit ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 77566.484 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: GenBank: AAH28384.1 / Source: (gene. exp.)  Homo sapiens (human) / Gene: GUCY1A1, GUC1A3, GUCSA3, GUCY1A3 / Production host: Homo sapiens (human) / Gene: GUCY1A1, GUC1A3, GUCSA3, GUCY1A3 / Production host:  |
|---|---|
| #2: Protein | Mass: 70599.320 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GUCY1B1, GUC1B3, GUCSB3, GUCY1B3 / Production host: Homo sapiens (human) / Gene: GUCY1B1, GUC1B3, GUCSB3, GUCY1B3 / Production host:  |
-Non-polymers , 4 types, 5 molecules 


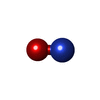



| #3: Chemical | | #4: Chemical | ChemComp-G2P / | #5: Chemical | ChemComp-HEM / | #6: Chemical | ChemComp-NO / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: human soluble guanylate cyclase / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 1500 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 QUANTUM (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20.1_4487: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 970628 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj












