+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7z5j | ||||||
|---|---|---|---|---|---|---|---|
| Title | The molybdenum storage protein loaded with tungstate | ||||||
 Components Components | (Molybdenum storage protein subunit ...) x 2 | ||||||
 Keywords Keywords | METAL BINDING PROTEIN / Metal storage protein / polytungstate / Keggin ion / EM | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.58 Å | ||||||
 Authors Authors | Ermler, U. / Aziz, I. / Kaltwasser, S. / Kayastha, K. / Vonck, J. | ||||||
| Funding support |  Germany, 1items Germany, 1items
| ||||||
 Citation Citation |  Journal: J Inorg Biochem / Year: 2022 Journal: J Inorg Biochem / Year: 2022Title: The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates. Authors: Iram Aziz / Susann Kaltwasser / Kanwal Kayastha / Radhika Khera / Janet Vonck / Ulrich Ermler /  Abstract: Some N-fixing bacteria store Mo to maintain the formation of the vital FeMo-cofactor dependent nitrogenase under Mo depleting conditions. The Mo storage protein (MoSto), developed for this purpose, ...Some N-fixing bacteria store Mo to maintain the formation of the vital FeMo-cofactor dependent nitrogenase under Mo depleting conditions. The Mo storage protein (MoSto), developed for this purpose, has the unique capability to compactly deposit molybdate as polyoxometalate (POM) clusters in a (αβ) hexameric cage; the same occurs with the physicochemically related tungstate. To explore the structural diversity of W-based POM clusters, MoSto loaded under different conditions with tungstate and two site-specifically modified MoSto variants were structurally characterized by X-ray crystallography or single-particle cryo-EM. The MoSto cage contains five major locations for POM clusters occupied among others by heptanuclear, Keggin ion and even Dawson-like species also found in bulk solvent under defined conditions. We found both lacunary derivatives of these archetypical POM clusters with missing WO units at positions exposed to bulk solvent and expanded derivatives with additional WO units next to protecting polypeptide segments or other POM clusters. The cryo-EM map, unexpectedly, reveals a POM cluster in the cage center anchored to the wall by a WO linker. Interestingly, distinct POM cluster structures can originate from identical, highly occupied core fragments of three to seven WO units that partly correspond to those found in MoSto loaded with molybdate. These core fragments are firmly bound to the complementary protein template in contrast to the more variable, less occupied residual parts of the visible POM clusters. Due to their higher stability, W-based POM clusters are, on average, larger and more diverse than their Mo-based counterparts. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7z5j.cif.gz 7z5j.cif.gz | 749.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7z5j.ent.gz pdb7z5j.ent.gz | 580 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7z5j.json.gz 7z5j.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/z5/7z5j https://data.pdbj.org/pub/pdb/validation_reports/z5/7z5j ftp://data.pdbj.org/pub/pdb/validation_reports/z5/7z5j ftp://data.pdbj.org/pub/pdb/validation_reports/z5/7z5j | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  14522MC  7zqqC  7zr4C  7zseC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Molybdenum storage protein subunit ... , 2 types, 12 molecules BDFHJLACEIGK
| #1: Protein | Mass: 28247.582 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azotobacter vinelandii DJ (bacteria) / Gene: mosB, Avin_43210 Azotobacter vinelandii DJ (bacteria) / Gene: mosB, Avin_43210Production host:  References: UniProt: P84253 #2: Protein | Mass: 29245.582 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Azotobacter vinelandii DJ (bacteria) / Strain: DJ / ATCC BAA-1303 / Gene: mosA, Avin_43200 Azotobacter vinelandii DJ (bacteria) / Strain: DJ / ATCC BAA-1303 / Gene: mosA, Avin_43200Production host:  References: UniProt: P84308 |
|---|
-Non-polymers , 9 types, 54 molecules 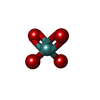


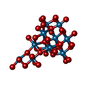
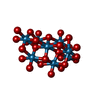

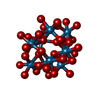









| #3: Chemical | ChemComp-MOO / #4: Chemical | ChemComp-ATP / #5: Chemical | ChemComp-MG / #6: Chemical | ChemComp-IWL / #7: Chemical | ChemComp-IV9 / |
|---|
 Movie
Movie Controller
Controller



 PDBj
PDBj


