Yorodumi
Yorodumi+ Open data
Open data
- Basic information
Basic information
| Entry | 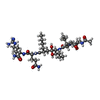 Database: PDB chemical components / ID: KVS Database: PDB chemical components / ID: KVS |
|---|---|
| Name | Name: |
-BIRD information
| Type | Peptide-like / Inhibitor |
|---|---|
 Downloads Downloads |  Molecular definition / Molecular definition /  Chemical definition / Chemical definition /  Family definition Family definition |
| Synonyms |
|
| Annotation |
|
| External info | |
| Family | ketomethylene isostere inhibitor N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: KVS / Ideal coordinates details: Corina / Model coordinates PDB-ID: 3NWX / Subcomponent: ACE, THR, ILE, NLH, GLN, ARG, NH2 | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  PubChem / PubChem /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 12.01 | | CACTVS 3.385 | OpenEye OEToolkits 1.7.6 | |
|---|
-SMILES CANONICAL
| CACTVS 3.385 | | OpenEye OEToolkits 1.7.6 | [ | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 12.01 | (| OpenEye OEToolkits 1.7.6 | ( | |
|---|
-PDB entries
Showing all 5 items

PDB-3dcr: 
X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor

PDB-3nwx: 
X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor

PDB-3nxn: 
X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor

PDB-6kmp: 
100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1

PDB-6ptp: 
Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1
 Movie
Movie Controller
Controller