+ Open data
Open data
- Basic information
Basic information
| Entry | 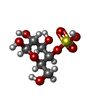 Database: PDB chemical components / ID: G4S Database: PDB chemical components / ID: G4S |
|---|---|
| Name | Name: Synonyms: 4-O-sulfo-beta-D-galactose; 4-O-sulfo-D-galactose; 4-O-sulfo-galactose |
-Chemical information
| Composition |  | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Others | Type: D-saccharide, beta linking / PDB classification: ATOMS / Three letter code: G4S / Ideal coordinates details: OpenEye/OEToolkits V1.4.2 / Model coordinates PDB-ID: 1CAR / Parent comp.: GAL / Replaces: GSA | ||||||||||||||
| History |
| ||||||||||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | [( | |
|---|
-CONDENSED IUPAC CARBOHYDRATE SYMBOL
| GMML 1.0 |
|---|
-COMMON NAME
| GMML 1.0 |
|---|
-IUPAC CARBOHYDRATE SYMBOL
| PDB-CARE 1.0 |
|---|
-PDB entries
Showing all 6 items

PDB-6b0j: 
Crystal structure of Ps i-CgsB in complex with k-i-k-neocarrahexaose

PDB-6b0k: 
Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose

PDB-6psm: 
Crystal structure of PsS1_19B C77S in complex with kappa-neocarrabiose

PDB-6pt9: 
Crystal structure of PsS1_NC C84S in complex with k-neocarrabiose

PDB-6ptk: 
Crystal structure of the sulfatase PsS1_NC C84A with bound sulfate ion

PDB-7oza: 
Sulfated host glycan recognition by carbohydrate sulfatases of the human gut microbiota (BT3796_S1_16)
 Movie
Movie Controller
Controller