[English] 日本語
 Yorodumi
Yorodumi- EMDB-9798: Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductois... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9798 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH7.5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bi-specific / Thermostable / Reductoisomerase / Magnesium-dependent / Dodecamer / Knot-structure / ISOMERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationketol-acid reductoisomerase (NADP+) / ketol-acid reductoisomerase activity / L-valine biosynthetic process / isoleucine biosynthetic process Similarity search - Function | |||||||||
| Biological species |   Sulfolobus solfataricus P2 (archaea) / Sulfolobus solfataricus P2 (archaea) /   Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea) Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Chen CY / Chang YC / Lin KF / Huang CH / Lin BL / Ko TP / Hsieh DL / Tsai MD | |||||||||
| Funding support |  Taiwan, 2 items Taiwan, 2 items
| |||||||||
 Citation Citation |  Journal: J Am Chem Soc / Year: 2019 Journal: J Am Chem Soc / Year: 2019Title: Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase. Authors: Chin-Yu Chen / Yuan-Chih Chang / Bo-Lin Lin / Kuan-Fu Lin / Chun-Hsiang Huang / Dong-Lin Hsieh / Tzu-Ping Ko / Ming-Daw Tsai /  Abstract: While cryo-EM is revolutionizing structural biology, its impact on enzymology is yet to be fully demonstrated. The ketol-acid reductoisomerase (KARI) catalyzes conversion of (2 S)-acetolactate or (2 ...While cryo-EM is revolutionizing structural biology, its impact on enzymology is yet to be fully demonstrated. The ketol-acid reductoisomerase (KARI) catalyzes conversion of (2 S)-acetolactate or (2 S)-aceto-2-hydroxybutyrate to 2,3-dihydroxy-3-alkylbutyrate. We found that KARI from archaea Sulfolobus solfataricus (Sso-KARI) is unusual in being a dodecamer, bispecific to NADH and NADPH, and losing activity above pH 7.8. While crystals were obtainable only at pH 8.5, cryo-EM structures were solved at pH 7.5 and 8.5 for Sso-KARI:2Mg. The results showed that the distances of the two catalytic Mg ions are lengthened in both structures at pH 8.5. We next solved cryo-EM structures of two Sso-KARI complexes, with NADH+inhibitor and NADPH+inhibitor at pH 7.5, which indicate that the bispecificity can be attributed to a unique asparagine at the cofactor binding loop. Unexpectedly, Sso-KARI also differs from other KARI enzymes in lacking "induced-fit", reflecting structural rigidity. Thus, cryo-EM is powerful for structural and mechanistic enzymology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9798.map.gz emd_9798.map.gz | 120 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9798-v30.xml emd-9798-v30.xml emd-9798.xml emd-9798.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9798.png emd_9798.png | 213.5 KB | ||
| Filedesc metadata |  emd-9798.cif.gz emd-9798.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9798 http://ftp.pdbj.org/pub/emdb/structures/EMD-9798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9798 | HTTPS FTP |
-Validation report
| Summary document |  emd_9798_validation.pdf.gz emd_9798_validation.pdf.gz | 536.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9798_full_validation.pdf.gz emd_9798_full_validation.pdf.gz | 536.1 KB | Display | |
| Data in XML |  emd_9798_validation.xml.gz emd_9798_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_9798_validation.cif.gz emd_9798_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9798 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9798 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9798 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9798 | HTTPS FTP |
-Related structure data
| Related structure data |  6jcvMC  9799C  9800C  9801C  6jcwC  6jczC  6jd1C  6jd2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9798.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9798.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
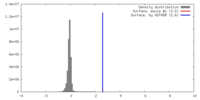
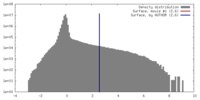
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) Do...
| Entire | Name: Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) Dodecamer in complex with Mg2+ in pH7.5 |
|---|---|
| Components |
|
-Supramolecule #1: Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) Do...
| Supramolecule | Name: Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) Dodecamer in complex with Mg2+ in pH7.5 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Sulfolobus solfataricus P2 (archaea) Sulfolobus solfataricus P2 (archaea) |
| Molecular weight | Theoretical: 446 kDa/nm |
-Macromolecule #1: Putative ketol-acid reductoisomerase 2
| Macromolecule | Name: Putative ketol-acid reductoisomerase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: ketol-acid reductoisomerase (NADP+) |
|---|---|
| Source (natural) | Organism:   Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea) Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea)Strain: ATCC 35092 / DSM 1617 / JCM 11322 / P2 |
| Molecular weight | Theoretical: 37.229855 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDKTVLDANL DPLKGKTIGV IGYGNQGRVQ ATIMRENGLN VIVGNVKDKY YELAKKEGFE VYEIDEAVRR SDVALLLIPD EVMKEVYEK KIAPVLQGKK EFVLDFASGY NVAFGLIRPP KSVDTIMVAP RMVGEGIMDL HKQGKGYPVL LGVKQDASGK A WDYAKAIA ...String: MDKTVLDANL DPLKGKTIGV IGYGNQGRVQ ATIMRENGLN VIVGNVKDKY YELAKKEGFE VYEIDEAVRR SDVALLLIPD EVMKEVYEK KIAPVLQGKK EFVLDFASGY NVAFGLIRPP KSVDTIMVAP RMVGEGIMDL HKQGKGYPVL LGVKQDASGK A WDYAKAIA KGIGAIPGGI AVISSFEEEA LLDLMSEHTW VPILFGAIKA CYDIAVKEYG VSPEAALLEF YASGELAEIA RL IAEEGIF NQMVHHSTTS QYGTLTRMFK YYDVVRRIVE NEAKYIWDGS FAKEWSLEQQ AGYPVFYRLW ELATQSEMAK AEK ELYKLL GRKVKND UniProtKB: Putative ketol-acid reductoisomerase 2 |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 24 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 96 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 1.91 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 123.03 |
|---|---|
| Output model |  PDB-6jcv: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)