+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8290 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GluK2EM with LY466195 | |||||||||
 Map data Map data | GluK2EM with LY466195 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GluK2EM with LY466195 / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity / inhibitory postsynaptic potential / glutamate receptor activity / ubiquitin conjugating enzyme binding ...mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity / inhibitory postsynaptic potential / glutamate receptor activity / ubiquitin conjugating enzyme binding / receptor clustering / modulation of excitatory postsynaptic potential / regulation of JNK cascade / kainate selective glutamate receptor activity / ionotropic glutamate receptor complex / extracellularly glutamate-gated ion channel activity / neuronal action potential / behavioral fear response / positive regulation of synaptic transmission / glutamate-gated receptor activity / glutamate-gated calcium ion channel activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / presynaptic modulation of chemical synaptic transmission / dendrite cytoplasm / hippocampal mossy fiber to CA3 synapse / regulation of membrane potential / SNARE binding / excitatory postsynaptic potential / synaptic transmission, glutamatergic / PDZ domain binding / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / modulation of chemical synaptic transmission / terminal bouton / intracellular calcium ion homeostasis / positive regulation of neuron apoptotic process / presynaptic membrane / scaffold protein binding / chemical synaptic transmission / perikaryon / postsynaptic membrane / neuron apoptotic process / negative regulation of neuron apoptotic process / postsynaptic density / axon / neuronal cell body / glutamatergic synapse / ubiquitin protein ligase binding / dendrite / synapse / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.6 Å | |||||||||
 Authors Authors | Meyerson JR / Chittori S | |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Structural basis of kainate subtype glutamate receptor desensitization. Authors: Joel R Meyerson / Sagar Chittori / Alan Merk / Prashant Rao / Tae Hee Han / Mihaela Serpe / Mark L Mayer / Sriram Subramaniam /  Abstract: Glutamate receptors are ligand-gated tetrameric ion channels that mediate synaptic transmission in the central nervous system. They are instrumental in vertebrate cognition and their dysfunction ...Glutamate receptors are ligand-gated tetrameric ion channels that mediate synaptic transmission in the central nervous system. They are instrumental in vertebrate cognition and their dysfunction underlies diverse diseases. In both the resting and desensitized states of AMPA and kainate receptor subtypes, the ion channels are closed, whereas the ligand-binding domains, which are physically coupled to the channels, adopt markedly different conformations. Without an atomic model for the desensitized state, it is not possible to address a central problem in receptor gating: how the resting and desensitized receptor states both display closed ion channels, although they have major differences in the quaternary structure of the ligand-binding domain. Here, by determining the structure of the kainate receptor GluK2 subtype in its desensitized state by cryo-electron microscopy (cryo-EM) at 3.8 Å resolution, we show that desensitization is characterized by the establishment of a ring-like structure in the ligand-binding domain layer of the receptor. Formation of this 'desensitization ring' is mediated by staggered helix contacts between adjacent subunits, which leads to a pseudo-four-fold symmetric arrangement of the ligand-binding domains, illustrating subtle changes in symmetry that are important for the gating mechanism. Disruption of the desensitization ring is probably the key switch that enables restoration of the receptor to its resting state, thereby completing the gating cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8290.map.gz emd_8290.map.gz | 96.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8290-v30.xml emd-8290-v30.xml emd-8290.xml emd-8290.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8290.png emd_8290.png | 46.7 KB | ||
| Filedesc metadata |  emd-8290.cif.gz emd-8290.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8290 http://ftp.pdbj.org/pub/emdb/structures/EMD-8290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8290 | HTTPS FTP |
-Validation report
| Summary document |  emd_8290_validation.pdf.gz emd_8290_validation.pdf.gz | 427.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8290_full_validation.pdf.gz emd_8290_full_validation.pdf.gz | 427.5 KB | Display | |
| Data in XML |  emd_8290_validation.xml.gz emd_8290_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_8290_validation.cif.gz emd_8290_validation.cif.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8290 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8290 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8290 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8290 | HTTPS FTP |
-Related structure data
| Related structure data |  5kuhMC  8289C  5cmkC  5cmmC  5kufC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8290.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8290.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK2EM with LY466195 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.324 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GluK2EM with LY466195
| Entire | Name: GluK2EM with LY466195 |
|---|---|
| Components |
|
-Supramolecule #1: GluK2EM with LY466195
| Supramolecule | Name: GluK2EM with LY466195 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor ionotropic, kainate 2
| Macromolecule | Name: Glutamate receptor ionotropic, kainate 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.526094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TTHVLRFGGI FEYVESGPMG AEELAFRFAV NTINRNRTLL PNTTLTYDTQ KINLYDSFEA SKKACDQLSL GVAAIFGPSH SSSANAVQS ICNALGVPHI QTRWKHQVSD NKDSFYVSLY PDFSSLSRAI LDLVQFFKWK TVTVVYDDST GLIRLQELIK A PSRYNLRL ...String: TTHVLRFGGI FEYVESGPMG AEELAFRFAV NTINRNRTLL PNTTLTYDTQ KINLYDSFEA SKKACDQLSL GVAAIFGPSH SSSANAVQS ICNALGVPHI QTRWKHQVSD NKDSFYVSLY PDFSSLSRAI LDLVQFFKWK TVTVVYDDST GLIRLQELIK A PSRYNLRL KIRQLPADTK DAKPLLKEMK RGKEFHVIFD CSHEMAAGIL KQALAMGMMT EYYHYIFTTL DLFALDVEPY RY SGVNMTG FRILNTENTQ VSSIIEKWSM ERLQAPPKPD SGLLDGFMTT DAALMYDAVH VVSVAVQQFP QMTVSSLQCN RHK PWRFGT RFMSLIKEAH WEGLTGRITF NKTNGLRTDF DLDVISLKEE GLEKIGTWDP ASGLNMTESQ KGKPANITDS LSNR SLIVT TILEEPYVLF KKSDKPLYGN DRFEGYCIDL LRELSTILGF TYEIRLVEDG KYGAQDDVNG QWNGMVRELI DHKAD LAVA PLTITYVREK VIDFSKPFMT LGISILYRKG TPIDSADDLA KQTKIEYGAV EDGSTMTFFK KSKISTYDKM WAFMSS RRQ SVLVKSSEEG IQRVLTSDYA LLMESTTIEF VTQRNCNLTQ IGGLIDSKGY GVGTPMGSPY RDKITIAILQ LQEEGKL HM MKEKWWRGNG CPEEESKEAS ALGVQNIGGI FIVLAAGLVL SVFVAVGEFL YKSKKNAQLE KRSFCSAMVE ELRMSLKC Q RRLKHKPQAP VIVKTEEVIN MHTFNDRRLP GKETMA UniProtKB: Glutamate receptor ionotropic, kainate 2, Glutamate receptor ionotropic, kainate 2 |
-Macromolecule #2: (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]me...
| Macromolecule | Name: (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid type: ligand / ID: 2 / Number of copies: 4 / Formula: LY5 |
|---|---|
| Molecular weight | Theoretical: 346.37 Da |
| Chemical component information | 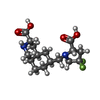 ChemComp-LY5: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
| Details | GluK2 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 11.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.3) / Number images used: 31000 |
| Initial angle assignment | Type: OTHER / Software - Name: RELION (ver. 1.3) |
| Final angle assignment | Type: OTHER / Software - Name: RELION (ver. 1.3) |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















