+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Active substate 4 of the GluA4 homotetramer. | ||||||||||||
 Map data Map data | Map corresponding to GluA4 active substate 4. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | AMPAR / Ion Channel / Glutamate Receptor / Ligand-gated / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationPresynaptic depolarization and calcium channel opening / LGI-ADAM interactions / Trafficking of AMPA receptors / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / regulation of AMPA receptor activity / Trafficking of AMPA receptors / kainate selective glutamate receptor complex ...Presynaptic depolarization and calcium channel opening / LGI-ADAM interactions / Trafficking of AMPA receptors / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / regulation of AMPA receptor activity / Trafficking of AMPA receptors / kainate selective glutamate receptor complex / membrane hyperpolarization / regulation of synapse structure or activity / nervous system process / Synaptic adhesion-like molecules / protein targeting to membrane / voltage-gated calcium channel complex / neurotransmitter receptor localization to postsynaptic specialization membrane / Activation of AMPA receptors / neuromuscular junction development / AMPA glutamate receptor activity / Trafficking of GluR2-containing AMPA receptors / negative regulation of smooth muscle cell apoptotic process / transmission of nerve impulse / AMPA glutamate receptor complex / ionotropic glutamate receptor complex / membrane depolarization / Unblocking of NMDA receptors, glutamate binding and activation / positive regulation of synaptic transmission, glutamatergic / regulation of postsynaptic membrane neurotransmitter receptor levels / response to fungicide / voltage-gated calcium channel activity / glutamate-gated receptor activity / presynaptic active zone membrane / somatodendritic compartment / ionotropic glutamate receptor binding / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / hippocampal mossy fiber to CA3 synapse / regulation of membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / synaptic transmission, glutamatergic / response to calcium ion / postsynaptic density membrane / modulation of chemical synaptic transmission / Schaffer collateral - CA1 synapse / terminal bouton / dendritic spine / chemical synaptic transmission / postsynaptic density / neuronal cell body / dendrite / synapse / glutamatergic synapse / cell surface / identical protein binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.89 Å | ||||||||||||
 Authors Authors | Hale WD / Huganir RL / Twomey EC | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Conformational Plasticity Tunes the GluA4 Channel Gate Authors: Hale WD / Huganir RL / Twomey EC | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_71409.map.gz emd_71409.map.gz | 122.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-71409-v30.xml emd-71409-v30.xml emd-71409.xml emd-71409.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_71409.png emd_71409.png | 64.3 KB | ||
| Filedesc metadata |  emd-71409.cif.gz emd-71409.cif.gz | 6.4 KB | ||
| Others |  emd_71409_half_map_1.map.gz emd_71409_half_map_1.map.gz emd_71409_half_map_2.map.gz emd_71409_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-71409 http://ftp.pdbj.org/pub/emdb/structures/EMD-71409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-71409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-71409 | HTTPS FTP |
-Related structure data
| Related structure data |  9p9fMC  9p9bC  9p9cC  9p9dC  9p9eC  9p9gC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_71409.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_71409.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map corresponding to GluA4 active substate 4. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A corresponding to GluA4 active substate 4.
| File | emd_71409_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A corresponding to GluA4 active substate 4. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B corresponding to GluA4 active substate 4.
| File | emd_71409_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B corresponding to GluA4 active substate 4. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octameric complex of four GluA4 subunits in complex with four TAR...
| Entire | Name: Octameric complex of four GluA4 subunits in complex with four TARP gamma 2 subunits. |
|---|---|
| Components |
|
-Supramolecule #1: Octameric complex of four GluA4 subunits in complex with four TAR...
| Supramolecule | Name: Octameric complex of four GluA4 subunits in complex with four TARP gamma 2 subunits. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Isoform 2 of Glutamate receptor 4
| Macromolecule | Name: Isoform 2 of Glutamate receptor 4 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.140234 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGKIMHISVL LSPVLWGLIF GVSSVQIGGL FIRNTDQEYT AFRLAIFLHN TSPNASEAPF NLVPHVDNIE TANSFAVTNA FCSQYSRGV FAIFGLYDKR SVHTLTSFCS ALHISLITPS FPTEGESQFV LQLRPSLRGA LLSLLDHYEW NCFVFLYDTD R GYSILQAI ...String: MGKIMHISVL LSPVLWGLIF GVSSVQIGGL FIRNTDQEYT AFRLAIFLHN TSPNASEAPF NLVPHVDNIE TANSFAVTNA FCSQYSRGV FAIFGLYDKR SVHTLTSFCS ALHISLITPS FPTEGESQFV LQLRPSLRGA LLSLLDHYEW NCFVFLYDTD R GYSILQAI MEKAGQNGWH VSAICVENFN DVSYRQLLEE LDRRQEKKFV IDCEIERLQN ILEQIVSVGK HVKGYHYIIA NL GFKDISL ERFIHGGANV TGFQLVDFNT PMVTKLMDRW KKLDQREYPG SETPPKYTSA LTYDGVLVMA ETFRSLRRQK IDI SRRGNA GDCLANPAAP WGQGIDMERT LKQVRIQGLT GNVQFDHYGR RVNYTMDVFE LKSTGPRKVG YWNDMDKLVL IQDM PTLGN DTAAIENRTV VVTTIMESPY VMYKKNHEMF EGNDKYEGYC VDLASEIAKH IGIKYKIAIV PDGKYGARDA DTKIW NGMV GELVYGKAEI AIAPLTITLV REEVIDFSKP FMSLGISIMI KKPQKSKPGV FSFLDPLAYE IWMCIVFAYI GVSVVL FLV SRFSPYEWHT EEPEDGKEGP SDQPPNEFGI FNSLWFSLGA FMQQGCDISP RSLSGRIVGG VWWFFTLIII SSYTANL AA FLTVERMVSP IESAEDLAKQ TEIAYGTLDS GSTKEFFRRS KIAVYEKMWT YMRSAEPSVF TRTTAEGVAR VRKSKGKF A FLLESTMNEY IEQRKPCDTM KVGGNLDSKG YGVATPKGSS LRTPVNLAVL KLSEAGVLDK LKNKWWYDKG ECGPKDSGS KDKTSALSLS NVAGVFYILV GGLGLAMLVA LIEFCYKSRA EAKRMK UniProtKB: Glutamate receptor 4 |
-Macromolecule #2: Voltage-dependent calcium channel gamma-2 subunit
| Macromolecule | Name: Voltage-dependent calcium channel gamma-2 subunit / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.066498 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLFDRGVQML LTTVGAFAAF SLMTIAVGTD YWLYSRGVCK TKSVSEDETS KKNEEVMTHS GLWRTCCLEG NFKGLCKQID HFPEDADYE ADTAEYFLRA VRASSIFPIL SVILLFMGGL CIAASEFYKT RHNIILSAGI FFVSAGLSNI IGIIVYISAN A GDPSKSDS ...String: GLFDRGVQML LTTVGAFAAF SLMTIAVGTD YWLYSRGVCK TKSVSEDETS KKNEEVMTHS GLWRTCCLEG NFKGLCKQID HFPEDADYE ADTAEYFLRA VRASSIFPIL SVILLFMGGL CIAASEFYKT RHNIILSAGI FFVSAGLSNI IGIIVYISAN A GDPSKSDS KKNSYSYGWS FYFGALSFII AEMVGVLAVH MFIDRHKQLT G UniProtKB: Voltage-dependent calcium channel gamma-2 subunit |
-Macromolecule #3: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 3 / Number of copies: 4 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Macromolecule #4: CYCLOTHIAZIDE
| Macromolecule | Name: CYCLOTHIAZIDE / type: ligand / ID: 4 / Number of copies: 4 / Formula: CYZ |
|---|---|
| Molecular weight | Theoretical: 389.878 Da |
| Chemical component information | 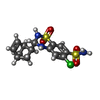 ChemComp-CYZ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































