+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PSS1 with calcium | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phosphatidylserine synthase / ER membrane protein / integral membrane protein / transmembrane enzyme / PS / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-serine-phosphatidylcholine phosphatidyltransferase activity / L-serine-phosphatidylethanolamine phosphatidyltransferase / L-serine-phosphatidylethanolamine phosphatidyltransferase activity / Synthesis of PS / phosphatidylserine biosynthetic process / transferase activity / endoplasmic reticulum membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Ning Y / Yu J / Ge J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2025 Journal: Cell Discov / Year: 2025Title: Structural basis for catalytic mechanism of human phosphatidylserine synthase 1. Authors: Yingjie Ning / Ruisheng Xu / Jie Yu / Jingpeng Ge /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62506.map.gz emd_62506.map.gz | 49.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62506-v30.xml emd-62506-v30.xml emd-62506.xml emd-62506.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_62506_fsc.xml emd_62506_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_62506.png emd_62506.png | 34.7 KB | ||
| Filedesc metadata |  emd-62506.cif.gz emd-62506.cif.gz | 5.8 KB | ||
| Others |  emd_62506_half_map_1.map.gz emd_62506_half_map_1.map.gz emd_62506_half_map_2.map.gz emd_62506_half_map_2.map.gz | 48.8 MB 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62506 http://ftp.pdbj.org/pub/emdb/structures/EMD-62506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62506 | HTTPS FTP |
-Related structure data
| Related structure data |  9kqjMC  9kqfC  9kqiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_62506.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62506.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||
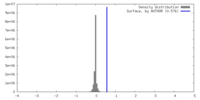
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_62506_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_62506_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : cryoEM structure of PSS1 dimer
| Entire | Name: cryoEM structure of PSS1 dimer |
|---|---|
| Components |
|
-Supramolecule #1: cryoEM structure of PSS1 dimer
| Supramolecule | Name: cryoEM structure of PSS1 dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylserine synthase 1
| Macromolecule | Name: Phosphatidylserine synthase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: L-serine-phosphatidylethanolamine phosphatidyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.590383 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASCVGSRTL SKDDVNYKMH FRMINEQQVE DITIDFFYRP HTITLLSFTI VSLMYFAFTR DDSVPEDNIW RGILSVIFFF LIISVLAFP NGPFTRPHPA LWRMVFGLSV LYFLFLVFLL FLNFEQVKSL MYWLDPNLRY ATREADVMEY AVNCHVITWE R IISHFDIF ...String: MASCVGSRTL SKDDVNYKMH FRMINEQQVE DITIDFFYRP HTITLLSFTI VSLMYFAFTR DDSVPEDNIW RGILSVIFFF LIISVLAFP NGPFTRPHPA LWRMVFGLSV LYFLFLVFLL FLNFEQVKSL MYWLDPNLRY ATREADVMEY AVNCHVITWE R IISHFDIF AFGHFWGWAM KALLIRSYGL CWTISITWEL TELFFMHLLP NFAECWWDQV ILDILLCNGG GIWLGMVVCR FL EMRTYHW ASFKDIHTTT GKIKRAVLQF TPASWTYVRW FDPKSSFQRV AGVYLFMIIW QLTELNTFFL KHIFVFQASH PLS WGRILF IGGITAPTVR QYYAYLTDTQ CKRVGTQCWV FGVIGFLEAI VCIKFGQDLF SKTQILYVVL WLLCVAFTTF LCLY GMIWY AEHYGHREKT YSECEDGTYS PEISWHHRKG TKGSEDSPPK HAGNNESHSS RRRNRHSKSK VTNGVGKK UniProtKB: Phosphatidylserine synthase 1 |
-Macromolecule #2: 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE
| Macromolecule | Name: 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE / type: ligand / ID: 2 / Number of copies: 4 / Formula: PSF |
|---|---|
| Molecular weight | Theoretical: 455.437 Da |
| Chemical component information |  ChemComp-PSF: |
-Macromolecule #3: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 3 / Number of copies: 10 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Macromolecule #4: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 4 / Number of copies: 2 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #5: TETRADECANE
| Macromolecule | Name: TETRADECANE / type: ligand / ID: 5 / Number of copies: 2 / Formula: C14 |
|---|---|
| Molecular weight | Theoretical: 198.388 Da |
| Chemical component information |  ChemComp-C14: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: Phosphatidylinositol
| Macromolecule | Name: Phosphatidylinositol / type: ligand / ID: 7 / Number of copies: 2 / Formula: T7X |
|---|---|
| Molecular weight | Theoretical: 887.128 Da |
| Chemical component information |  ChemComp-T7X: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)