+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | the wild-type human PRMT1 filament | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protein Arginine Methyltransferase / filament / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationGATOR1 complex binding / positive regulation of hemoglobin biosynthetic process / protein-arginine omega-N monomethyltransferase activity / N-methyltransferase activity / peptidyl-arginine methylation / regulation of BMP signaling pathway / protein-arginine omega-N asymmetric methyltransferase activity / regulation of megakaryocyte differentiation / type I protein arginine methyltransferase / histone H4R3 methyltransferase activity ...GATOR1 complex binding / positive regulation of hemoglobin biosynthetic process / protein-arginine omega-N monomethyltransferase activity / N-methyltransferase activity / peptidyl-arginine methylation / regulation of BMP signaling pathway / protein-arginine omega-N asymmetric methyltransferase activity / regulation of megakaryocyte differentiation / type I protein arginine methyltransferase / histone H4R3 methyltransferase activity / protein methyltransferase activity / protein methylation / protein-arginine N-methyltransferase activity / cellular response to methionine / methylosome / S-adenosyl-L-methionine binding / methyl-CpG binding / positive regulation of p38MAPK cascade / negative regulation of JNK cascade / cardiac muscle tissue development / Maturation of nucleoprotein / mitogen-activated protein kinase p38 binding / histone methyltransferase activity / positive regulation of double-strand break repair via homologous recombination / negative regulation of megakaryocyte differentiation / positive regulation of TORC1 signaling / RNA splicing / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / positive regulation of erythrocyte differentiation / positive regulation of translation / methyltransferase activity / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / protein homooligomerization / RMTs methylate histone arginines / neuron projection development / Estrogen-dependent gene expression / in utero embryonic development / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / viral protein processing / chromatin remodeling / lysosomal membrane / positive regulation of cell population proliferation / DNA damage response / regulation of DNA-templated transcription / enzyme binding / RNA binding / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
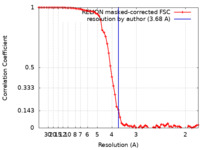
| Method | single particle reconstruction / cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Li YW / Ru YX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the human PRMT1 filament Authors: Li YW / Ru YX | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61691.map.gz emd_61691.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61691-v30.xml emd-61691-v30.xml emd-61691.xml emd-61691.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61691_fsc.xml emd_61691_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_61691.png emd_61691.png | 105.6 KB | ||
| Filedesc metadata |  emd-61691.cif.gz emd-61691.cif.gz | 5.4 KB | ||
| Others |  emd_61691_half_map_1.map.gz emd_61691_half_map_1.map.gz emd_61691_half_map_2.map.gz emd_61691_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61691 http://ftp.pdbj.org/pub/emdb/structures/EMD-61691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61691 | HTTPS FTP |
-Related structure data
| Related structure data |  9jp0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61691.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61691.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: 2
| File | emd_61691_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 1
| File | emd_61691_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : the wild type PRMT1
| Entire | Name: the wild type PRMT1 |
|---|---|
| Components |
|
-Supramolecule #1: the wild type PRMT1
| Supramolecule | Name: the wild type PRMT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein arginine N-methyltransferase 1
| Macromolecule | Name: Protein arginine N-methyltransferase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO / EC number: type I protein arginine methyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.511551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAAEAANCI MENFVATLAN GMSLQPPLEE VSCGQAESSE KPNAEDMTSK DYYFDSYAHF GIHEEMLKDE VRTLTYRNSM FHNRHLFKD KVVLDVGSGT GILCMFAAKA GARKVIGIEC SSISDYAVKI VKANKLDHVV TIIKGKVEEV ELPVEKVDII I SEWMGYCL ...String: MAAAEAANCI MENFVATLAN GMSLQPPLEE VSCGQAESSE KPNAEDMTSK DYYFDSYAHF GIHEEMLKDE VRTLTYRNSM FHNRHLFKD KVVLDVGSGT GILCMFAAKA GARKVIGIEC SSISDYAVKI VKANKLDHVV TIIKGKVEEV ELPVEKVDII I SEWMGYCL FYESMLNTVL YARDKWLAPD GLIFPDRATL YVTAIEDRQY KDYKIHWWEN VYGFDMSCIK DVAIKEPLVD VV DPKQLVT NACLIKEVDI YTVKVEDLTF TSPFCLQVKR NDYVHALVAY FNIEFTRCHK RTGFSTSPES PYTHWKQTVF YME DYLTVK TGEEIFGTIG MRPNAKNNRD LDFTIDLDFK GQLCELSCST DYRMR UniProtKB: Protein arginine N-methyltransferase 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)