+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the C18:0 ceramide-bound FPR2-Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / ceramide / Membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationN-formyl peptide receptor activity / complement receptor activity / immune response-regulating cell surface receptor signaling pathway / scavenger receptor binding / RAGE receptor binding / complement receptor mediated signaling pathway / positive regulation of monocyte chemotaxis / Formyl peptide receptors bind formyl peptides and many other ligands / positive regulation of innate immune response / cargo receptor activity ...N-formyl peptide receptor activity / complement receptor activity / immune response-regulating cell surface receptor signaling pathway / scavenger receptor binding / RAGE receptor binding / complement receptor mediated signaling pathway / positive regulation of monocyte chemotaxis / Formyl peptide receptors bind formyl peptides and many other ligands / positive regulation of innate immune response / cargo receptor activity / positive chemotaxis / tertiary granule membrane / ficolin-1-rich granule membrane / specific granule membrane / adenylate cyclase inhibitor activity / positive regulation of superoxide anion generation / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / astrocyte activation / receptor-mediated endocytosis / cellular response to forskolin / positive regulation of phagocytosis / regulation of mitotic spindle organization / Regulation of insulin secretion / microglial cell activation / calcium-mediated signaling / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / negative regulation of inflammatory response / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / cellular response to amyloid-beta / Olfactory Signaling Pathway / chemotaxis / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / signaling receptor activity / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / amyloid-beta binding / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / negative regulation of neuron apoptotic process Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Sun JP / Jiang CT / Kong W / Yu X / Cai K / Guo LL | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2025 Journal: Science / Year: 2025Title: Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis. Authors: Hui Lin / Chuanshun Ma / Kui Cai / Lulu Guo / Xuemei Wang / Lin Lv / Chao Zhang / Jun Lin / Daolai Zhang / Chuan Ye / Tengwei Wang / Shenming Huang / Jifei Han / Zihao Zhang / Junyan Gao / ...Authors: Hui Lin / Chuanshun Ma / Kui Cai / Lulu Guo / Xuemei Wang / Lin Lv / Chao Zhang / Jun Lin / Daolai Zhang / Chuan Ye / Tengwei Wang / Shenming Huang / Jifei Han / Zihao Zhang / Junyan Gao / Mingxiang Zhang / Zhao Pu / Fengyang Li / Yongyuan Guo / Xiaojun Zhou / Chengxue Qin / Fan Yi / Xiao Yu / Wei Kong / Changtao Jiang / Jin-Peng Sun /   Abstract: Ceramides play a central role in human health and disease, yet their role as systemic signaling molecules remain poorly understood. In this work, we identify formyl peptide receptor 2 (FPR2) as a ...Ceramides play a central role in human health and disease, yet their role as systemic signaling molecules remain poorly understood. In this work, we identify formyl peptide receptor 2 (FPR2) as a membrane receptor that specifically binds long-chain ceramides (C14 to C20). In brown and beige adipocytes, C16:0 ceramide binding to FPR2 inhibits thermogenesis through G cyclic adenosine monophosphate signaling pathways, an effect that is reversed in the absence of FPR2. We present three cryo-electron microscopy structures of FPR2 in complex with G trimers bound to C16:0, C18:0, and C20:0 ceramides. The hydrophobic tails are deeply embedded in the orthosteric ligand pocket, which has a limited amount of plasticity. Modification of the ceramide binding motif in closely related receptors, such as FPR1 or FPR3, converts them from inactive to active ceramide receptors. Our findings provide a structural basis for adipocyte thermogenesis mediated by FPR2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61484.map.gz emd_61484.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61484-v30.xml emd-61484-v30.xml emd-61484.xml emd-61484.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61484_fsc.xml emd_61484_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_61484.png emd_61484.png | 100.6 KB | ||
| Filedesc metadata |  emd-61484.cif.gz emd-61484.cif.gz | 7.4 KB | ||
| Others |  emd_61484_half_map_1.map.gz emd_61484_half_map_1.map.gz emd_61484_half_map_2.map.gz emd_61484_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61484 http://ftp.pdbj.org/pub/emdb/structures/EMD-61484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61484 | HTTPS FTP |
-Related structure data
| Related structure data |  9jhjMC  8y62C  8y63C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61484.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61484.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61484_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61484_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the C18:0 ceramide bound FPR2-Gi complex
| Entire | Name: Cryo-EM structure of the C18:0 ceramide bound FPR2-Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the C18:0 ceramide bound FPR2-Gi complex
| Supramolecule | Name: Cryo-EM structure of the C18:0 ceramide bound FPR2-Gi complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: N-formyl peptide receptor 2
| Macromolecule | Name: N-formyl peptide receptor 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.993055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METNFSTPLN EYEEVSYESA GYTVLRILPL VVLGVTFVLG VLGNGLVIWV AGFRMTRTVT TICYLNLALA DFSFTATLPF LIVSMAMGE KWPFGWFLCK LIHIVVDINL FGSVFLIGFI ALDRCICVLH PVWAQNHRTV SLAMKVIVGP WILALVLTLP V FLFLTTVT ...String: METNFSTPLN EYEEVSYESA GYTVLRILPL VVLGVTFVLG VLGNGLVIWV AGFRMTRTVT TICYLNLALA DFSFTATLPF LIVSMAMGE KWPFGWFLCK LIHIVVDINL FGSVFLIGFI ALDRCICVLH PVWAQNHRTV SLAMKVIVGP WILALVLTLP V FLFLTTVT IPNGDTYCTF NFASWGGTPE ERLKVAITML TARGIIRFVI GFSLPMSIVA ICYGLIAAKI HKKGMIKSSR PL RVLTAVV ASFFICWFPF QLVALLGTVW LKEMLFYGKY KIIDILVNPT SSLAFFNSCL NPMLYVFVGQ DFRERLIHSL PTS LERALS EDSAPTNDTA ANSASPPAET ELQAM UniProtKB: N-formyl peptide receptor 2 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.363043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSADIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLEL |
-Macromolecule #6: N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE
| Macromolecule | Name: N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE type: ligand / ID: 6 / Number of copies: 1 / Formula: 18C |
|---|---|
| Molecular weight | Theoretical: 565.954 Da |
| Chemical component information | 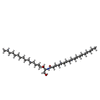 ChemComp-18C: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Average electron dose: 1.875 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)