+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Epinephrine-activated human beta3 adrenergic receptor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / beta3AR / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationbeta-3 adrenergic receptor binding / beta3-adrenergic receptor activity / beta-adrenergic receptor activity / epinephrine binding / sensory perception of chemical stimulus / energy reserve metabolic process / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / G-protein activation / Activation of the phototransduction cascade ...beta-3 adrenergic receptor binding / beta3-adrenergic receptor activity / beta-adrenergic receptor activity / epinephrine binding / sensory perception of chemical stimulus / energy reserve metabolic process / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / heat generation / norepinephrine binding / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Adrenoceptors / G alpha (q) signalling events / beta-2 adrenergic receptor binding / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / eating behavior / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / negative regulation of multicellular organism growth / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / diet induced thermogenesis / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / photoreceptor outer segment / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / D1 dopamine receptor binding / brown fat cell differentiation / adenylate cyclase-activating adrenergic receptor signaling pathway / insulin-like growth factor receptor binding / cardiac muscle cell apoptotic process / photoreceptor inner segment / ionotropic glutamate receptor binding / response to cold / adenylate cyclase activator activity / generation of precursor metabolites and energy / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / sensory perception of taste / positive regulation of cold-induced thermogenesis / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / cell body / GTPase binding / cellular response to hypoxia / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / carbohydrate metabolic process / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / receptor complex / cell population proliferation / positive regulation of MAPK cascade Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.34 Å | |||||||||
 Authors Authors | Zheng S / Zhang S / Dai S / Chen K / Gao K / Lin B / Liu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Chempluschem / Year: 2024 Journal: Chempluschem / Year: 2024Title: Molecular Mechanism of the βAR Agonist Activity of a β-Blocker. Authors: Shuang Zheng / Shuhao Zhang / Shengjie Dai / Kai Chen / Kaixuan Gao / Xiaoou Sun / Bin Lin / Xiangyu Liu /  Abstract: Development of subtype-selective drugs for G protein-coupled receptors poses a significant challenge due to high similarity between subtypes, as exemplified by the three β-adrenergic receptors ...Development of subtype-selective drugs for G protein-coupled receptors poses a significant challenge due to high similarity between subtypes, as exemplified by the three β-adrenergic receptors (βARs). The βAR agonists show promise for treating the overactive bladder or preterm birth, but their potential is hindered by off-target activation of βAR and βAR. Interestingly, several β-blockers, which are antagonists of the βARs and βARs, have been reported to exhibit agonist activity at the βAR. However, the molecular mechanism remains elusive. Understanding the underlying mechanism should facilitate the development of βAR agonists with improved selectivity and reduced off-target effects. In this work, we determined the structures of human βAR in complex with the endogenous agonist epinephrine or with a synthetic βAR agonist carazolol, which is also a high-affinity β-blocker. Structure comparison, mutagenesis studies and molecular dynamics simulations revealed that the differences on the flexibility of D directly contribute to carazolol's distinct activities as an antagonist for the βAR and an agonist for the βAR. The process is also indirectly influenced by the extracellular loops (ECL), especially ECL1. Taken together, these results provide key guidance for development of selective βAR agonists, paving the way for new therapeutic opportunities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60629.map.gz emd_60629.map.gz | 42.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60629-v30.xml emd-60629-v30.xml emd-60629.xml emd-60629.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60629.png emd_60629.png | 87.7 KB | ||
| Filedesc metadata |  emd-60629.cif.gz emd-60629.cif.gz | 7.3 KB | ||
| Others |  emd_60629_half_map_1.map.gz emd_60629_half_map_1.map.gz emd_60629_half_map_2.map.gz emd_60629_half_map_2.map.gz | 42 MB 42 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60629 http://ftp.pdbj.org/pub/emdb/structures/EMD-60629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60629 | HTTPS FTP |
-Related structure data
| Related structure data |  9ijeMC  9ijdC  39845 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60629.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60629.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
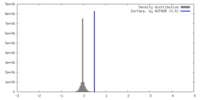
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60629_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
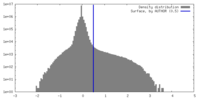
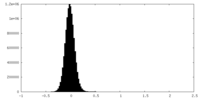
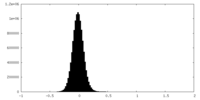
| Density Histograms |
-Half map: #1
| File | emd_60629_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
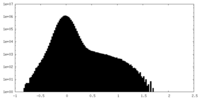
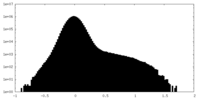
| Density Histograms |
- Sample components
Sample components
-Entire : Epinephrine bounded human beta3 adrenergic receptor-Gs protein co...
| Entire | Name: Epinephrine bounded human beta3 adrenergic receptor-Gs protein complex with Gbeta, Ggamma, Nb35 and scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: Epinephrine bounded human beta3 adrenergic receptor-Gs protein co...
| Supramolecule | Name: Epinephrine bounded human beta3 adrenergic receptor-Gs protein complex with Gbeta, Ggamma, Nb35 and scFv16 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.413863 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: QSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: QSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #2: Camelid antibody VHH fragment
| Macromolecule | Name: Camelid antibody VHH fragment / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.885439 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSS |
-Macromolecule #3: Single-chain Fv16
| Macromolecule | Name: Single-chain Fv16 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.206219 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS SGGGGSGGGG SGGGGADIVM TQATSSVPVT PGESVSISCR S SKSLLHSN ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS SGGGGSGGGG SGGGGADIVM TQATSSVPVT PGESVSISCR S SKSLLHSN GNTYLYWFLQ RPGQSPQLLI YRMSNLASGV PDRFSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GA GTKLEL |
-Macromolecule #4: Beta-3 adrenergic receptor
| Macromolecule | Name: Beta-3 adrenergic receptor / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.64702 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ALLALAVLAT VGGNLLVIVA IAWTPRLQTM TNVFVTSLAA ADLVMGLLVV PPAATLALTG HWPLGATGCE LWTSVDVLCV TASIWTLCA LAVDRYLAVT NPLRYGALVT KRCARTAVVL VWVVSAAVSF APIMSQWWRV GADAEAQRCH SNPRCCAFAS N MPYVLLSS ...String: ALLALAVLAT VGGNLLVIVA IAWTPRLQTM TNVFVTSLAA ADLVMGLLVV PPAATLALTG HWPLGATGCE LWTSVDVLCV TASIWTLCA LAVDRYLAVT NPLRYGALVT KRCARTAVVL VWVVSAAVSF APIMSQWWRV GADAEAQRCH SNPRCCAFAS N MPYVLLSS SVSFYLPLLV MLFVYARVFV VATRQLRLLR GELGRFPPEE SPPAPSRSLA PAPVGTCAPP EGVPACGRRP AR LLPLREH RALCTLGLIM GTFTLCWLPF FLANVLRALG GPSLVPGPAF LALNWLGYAN SAFNPLIYCR SPDFRSAFRR LLC R UniProtKB: Beta-3 adrenergic receptor |
-Macromolecule #5: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.320797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LSAEDKAAVE RSKMIEKQLQ KDKQVYRATH RLLLLGADNS GKSTIVKQMR ILHVNGFNGE GGEEDPQAAR SNSDGEKATK VQDIKNNLK EAIETIVAAM SNLVPPVELA NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR ACYERSNEYQ L IDCAQYFL ...String: LSAEDKAAVE RSKMIEKQLQ KDKQVYRATH RLLLLGADNS GKSTIVKQMR ILHVNGFNGE GGEEDPQAAR SNSDGEKATK VQDIKNNLK EAIETIVAAM SNLVPPVELA NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR ACYERSNEYQ L IDCAQYFL DKIDVIKQDD YVPSDQDLLR CRVLTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DVTAIIFVVD SS DYNRLQE ALNLFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI EDYFPEFARY TTPEDATPEP GEDPRVTRAK YFI RDEFLR ISTASGDGRH YCYPHFTCAV DTENARRIFN DCRDIIQRMH LRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #6: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.731619 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: AQARKLVEQL KMEANIDRIK VSKAAADLMA YCEAHAKEDP LLTPVPASEN PF UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #7: L-EPINEPHRINE
| Macromolecule | Name: L-EPINEPHRINE / type: ligand / ID: 7 / Number of copies: 1 / Formula: ALE |
|---|---|
| Molecular weight | Theoretical: 183.204 Da |
| Chemical component information |  ChemComp-ALE: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.1 µm |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)