[English] 日本語
 Yorodumi
Yorodumi- EMDB-53173: Cryo-EM structure of mouse TRPM3 alpha 2 in complex with antagoni... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse TRPM3 alpha 2 in complex with antagonist Ononetin | |||||||||
 Map data Map data | Cryo-EM structure of mouse TRPM3 alpha 2 in complex with antagonist Ononetin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ca2+ channel Ononetin-bound Closed conformation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein tetramerization / calcium channel activity / calmodulin binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Shkumatov AV / Schenck S / Brunner JD | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of mouse TRPM3 alpha 2 in complex with antagonist Ononetin Authors: Shkumatov AV / Schenck S / Brunner JD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53173.map.gz emd_53173.map.gz | 237.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53173-v30.xml emd-53173-v30.xml emd-53173.xml emd-53173.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_53173_fsc.xml emd_53173_fsc.xml | 18.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_53173.png emd_53173.png | 128.8 KB | ||
| Masks |  emd_53173_msk_1.map emd_53173_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-53173.cif.gz emd-53173.cif.gz | 7.5 KB | ||
| Others |  emd_53173_additional_1.map.gz emd_53173_additional_1.map.gz emd_53173_additional_2.map.gz emd_53173_additional_2.map.gz emd_53173_half_map_1.map.gz emd_53173_half_map_1.map.gz emd_53173_half_map_2.map.gz emd_53173_half_map_2.map.gz | 237.7 MB 104 MB 441.4 MB 441.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53173 http://ftp.pdbj.org/pub/emdb/structures/EMD-53173 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53173 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53173 | HTTPS FTP |
-Related structure data
| Related structure data |  9qhmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_53173.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53173.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of mouse TRPM3 alpha 2 in complex with antagonist Ononetin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.693 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_53173_msk_1.map emd_53173_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Input for consensus map together with main map
| File | emd_53173_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Input for consensus map together with main map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Consensus map used for model building and phenix validation
| File | emd_53173_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map used for model building and phenix validation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfB
| File | emd_53173_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfA
| File | emd_53173_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric assembly of mouse TRPM3 alpha 2 with inhibitor Ononetin
| Entire | Name: Tetrameric assembly of mouse TRPM3 alpha 2 with inhibitor Ononetin |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric assembly of mouse TRPM3 alpha 2 with inhibitor Ononetin
| Supramolecule | Name: Tetrameric assembly of mouse TRPM3 alpha 2 with inhibitor Ononetin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 161 KDa |
-Macromolecule #1: MKIAA1616 protein
| Macromolecule | Name: MKIAA1616 protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 161.96325 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSGKKWRDAG ELERGCSDRE DSAESRRRSR SASRGRFAES WKRLSSKQGS TKRSGLPAQQ TPAQKSWIER AFYKRECVHI IPSTKDPHR CCCGRLIGQH VGLTPSISVL QNEKNESRLS RNDIQSEKWS ISKHTQLSPT DAFGTIEFQG GGHSNKAMYV R VSFDTKPD ...String: MSGKKWRDAG ELERGCSDRE DSAESRRRSR SASRGRFAES WKRLSSKQGS TKRSGLPAQQ TPAQKSWIER AFYKRECVHI IPSTKDPHR CCCGRLIGQH VGLTPSISVL QNEKNESRLS RNDIQSEKWS ISKHTQLSPT DAFGTIEFQG GGHSNKAMYV R VSFDTKPD LLLHLMTKEW QLELPKLLIS VHGGLQNFEL QPKLKQVFGK GLIKAAMTTG AWIFTGGVNT GVIRHVGDAL KD HASKSRG KICTIGIAPW GIVENQEDLI GRDVVRPYQT MSNPMSKLTV LNSMHSHFIL ADNGTTGKYG AEVKLRRQLE KHI SLQKIN TRIGQGVPVV ALIVEGGPNV ISIVLEYLRD TPPVPVVVCD GSGRASDILA FGHKYSEEGG LINESLRDQL LVTI QKTFT YTRTQAQHLF IILMECMKKK ELITVFRMGS EGHQDIDLAI LTALLKGANA SAPDQLSLAL AWNRVDIARS QIFIY GQQW PVGSLEQAML DALVLDRVDF VKLLIENGVS MHRFLTISRL EELYNTRHGP SNTLYHLVRD VKKGNLPPDY RISLID IGL VIEYLMGGAY RCNYTRKRFR TLYHNLFGPK RPKALKLLGM EDDIPLRRGR KTTKKREEEV DIDLDDPEIN HFPFPFH EL MVWAVLMKRQ KMALFFWQHG EEAMAKALVA CKLCKAMAHE ASENDMVDDI SQELNHNSRD FGQLAVELLD QSYKQDEQ L AMKLLTYELK NWSNATCLQL AVAAKHRDFI AHTCSQMLLT DMWMGRLRMR KNSGLKVILG ILLPPSILSL EFKNKDDMP YMTQAQEIHL QEKEPEEPEK PTKEKDEEDM ELTAMLGRSN GESSRKKDEE EVQSRHRLIP VGRKIYEFYN APIVKFWFYT LAYIGYLML FNYIVLVKME RWPSTQEWIV ISYIFTLGIE KMREILMSEP GKLLQKVKVW LQEYWNVTDL IAILLFSVGM I LRLQDQPF RSDGRVIYCV NIIYWYIRLL DIFGVNKYLG PYVMMIGKMM IDMMYFVIIM LVVLMSFGVA RQAILFPNEE PS WKLAKNI FYMPYWMIYG EVFADQIDPP CGQNETREDG KTIQLPPCKT GAWIVPAIMA CYLLVANILL VNLLIAVFNN TFF EVKSIS NQVWKFQRYQ LIMTFHERPV LPPPLIIFSH MTMIFQHVCC RWRKHESDQD ERDYGLKLFI TDDELKKVHD FEEQ CIEEY FREKDDRFNS SNDERIRVTS ERVENMSMRL EEVNEREHSM KASLQTVDIR LAQLEDLIGR MATALERLTG LERAE SNKI RSRTSSDCTD AAYIVRQSSF NSQEGNTFKL QESIDPAGEE TISPTSPTLM PRMRSHSFYS VALEVLFQGP QGTEQK LIS EEDLRGASMD EKTTGWRGGH VVEGLAGELE QLRARLEHHP QGQREP UniProtKB: Transient receptor potential cation channel subfamily M member 3 |
-Macromolecule #2: 1-[2,4-bis(oxidanyl)phenyl]-2-(4-methoxyphenyl)ethanone
| Macromolecule | Name: 1-[2,4-bis(oxidanyl)phenyl]-2-(4-methoxyphenyl)ethanone type: ligand / ID: 2 / Number of copies: 4 / Formula: A1D6M |
|---|---|
| Molecular weight | Theoretical: 258.269 Da |
-Macromolecule #3: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
| Macromolecule | Name: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: 3PH |
|---|---|
| Molecular weight | Theoretical: 704.998 Da |
| Chemical component information |  ChemComp-3PH: |
-Macromolecule #4: (25R)-14beta,17beta-spirost-5-en-3beta-ol
| Macromolecule | Name: (25R)-14beta,17beta-spirost-5-en-3beta-ol / type: ligand / ID: 4 / Number of copies: 8 / Formula: YUV |
|---|---|
| Molecular weight | Theoretical: 414.621 Da |
| Chemical component information | 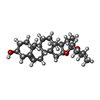 ChemComp-YUV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 10 mM Hepes pH 7.5, 150 mM NaCl, 0.063% Glycodiosgenin, 10 uM Ononetin | |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 279.15 K / Instrument: LEICA EM GP / Details: GP2. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.7000000000000001 µm |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






























































