+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BetP heterotrimeric complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycine betaine transporter BetP / TRANSPORT PROTEIN | |||||||||
| Function / homology | BCCT transporter, conserved site / BCCT family of transporters signature. / BCCT transporter family / BCCT, betaine/carnitine/choline family transporter / symporter activity / metal ion binding / identical protein binding / plasma membrane / Glycine betaine transporter BetP Function and homology information Function and homology information | |||||||||
| Biological species |  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Urbansky K / Fu L / Madej MG / Ziegler C | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Structure of Glycine betaine transporter BetP heterotrimeric complex Authors: Urbansky K / Fu L / Madej MG / Ziegler C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51422.map.gz emd_51422.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51422-v30.xml emd-51422-v30.xml emd-51422.xml emd-51422.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51422_fsc.xml emd_51422_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_51422.png emd_51422.png | 123.8 KB | ||
| Filedesc metadata |  emd-51422.cif.gz emd-51422.cif.gz | 7.2 KB | ||
| Others |  emd_51422_half_map_1.map.gz emd_51422_half_map_1.map.gz emd_51422_half_map_2.map.gz emd_51422_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51422 http://ftp.pdbj.org/pub/emdb/structures/EMD-51422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51422 | HTTPS FTP |
-Validation report
| Summary document |  emd_51422_validation.pdf.gz emd_51422_validation.pdf.gz | 902.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_51422_full_validation.pdf.gz emd_51422_full_validation.pdf.gz | 902.1 KB | Display | |
| Data in XML |  emd_51422_validation.xml.gz emd_51422_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_51422_validation.cif.gz emd_51422_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51422 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51422 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51422 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51422 | HTTPS FTP |
-Related structure data
| Related structure data |  9gkkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_51422.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51422.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7891 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_51422_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_51422_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Glycine Betaine Transporter BetP Heterotrimer
| Entire | Name: Glycine Betaine Transporter BetP Heterotrimer |
|---|---|
| Components |
|
-Supramolecule #1: Glycine Betaine Transporter BetP Heterotrimer
| Supramolecule | Name: Glycine Betaine Transporter BetP Heterotrimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
-Macromolecule #1: Glycine betaine transporter BetP
| Macromolecule | Name: Glycine betaine transporter BetP / type: protein_or_peptide / ID: 1 Details: >chainA: N-terminal StrepII, R584C WSHPQFEKMTTSDPNPKPIVEDAQPEQITATEELAGLLENPTNLEGKLADAEEEIILEGEDTQASLNW SVIVPALVIVLATVVWGIGFKDSFTNFASSALSAVVDNLGWAFILFGTVFVFFIVVIAAS ...Details: >chainA: N-terminal StrepII, R584C WSHPQFEKMTTSDPNPKPIVEDAQPEQITATEELAGLLENPTNLEGKLADAEEEIILEGEDTQASLNW SVIVPALVIVLATVVWGIGFKDSFTNFASSALSAVVDNLGWAFILFGTVFVFFIVVIAAS KFGTIRLGRIDEAPEFRTVSWISMMFAAGMGIGLMFYGTTEPLTFYRNGVPGHDEHNVGV AMSTTMFHWTLHPWAIYAIVGLAIAYSTFRVGRKQLLSSAFVPLIGEKGAEGWLGKLIDI LAIIATVFGTACSLGLGALQIGAGLSAANIIEDPSDWTIVGIVSVLTLAFIFSAISGVGK GIQYLSNANMVLAALLAIFVFVVGPTVSILNLLPGSIGNYLSNFFQMAGRTAMSADGTAG EWLGSWTIFYWAWWISWSPFVGMFLARISRGRSIREFILGVLLVPAGVSTVWFSIFGGTA IVFEQNGESIWGDGAAEEQLFGLLHALPGGQIMGIIAMILLGTFFITSADSASTVMGTMS QHGQLEANKWVTAAWGVATAAIGLTLLLSGGDNALSNLQNVTIVAATPFLFVVIGLMFAL VKDLSNDVIYLEYREQQRFNARLARERRVHNEHRKRELAAKRRCERKASGAGKRR >chainB: N-terminal StrepII, delC12, C-terminal His WSHPQFEKMTTSDPNPKPIVEDAQPEQITATEELAGLLENPTNLEGKLADAEEEIILEGEDTQASLNW SVIVPALVIVLATVVWGIGFKDSFTNFASSALSAVVDNLGWAFILFGTVFVFFIVVIAAS KFGTIRLGRIDEAPEFRTVSWISMMFAAGMGIGLMFYGTTEPLTFYRNGVPGHDEHNVGV AMSTTMFHWTLHPWAIYAIVGLAIAYSTFRVGRKQLLSSAFVPLIGEKGAEGWLGKLIDI LAIIATVFGTACSLGLGALQIGAGLSAANIIEDPSDWTIVGIVSVLTLAFIFSAISGVGK GIQYLSNANMVLAALLAIFVFVVGPTVSILNLLPGSIGNYLSNFFQMAGRTAMSADGTAG EWLGSWTIFYWAWWISWSPFVGMFLARISRGRSIREFILGVLLVPAGVSTVWFSIFGGTA IVFEQNGESIWGDGAAEEQLFGLLHALPGGQIMGIIAMILLGTFFITSADSASTVMGTMS QHGQLEANKWVTAAWGVATAAIGLTLLLSGGDNALSNLQNVTIVAATPFLFVVIGLMFAL VKDLSNDVIYLEYREQQRFNARLARERRVHNEHRKRELAAKRRHHHHHH >chainC: N-terminal StrepII, E25C, delC12, C-terminal Flag WSHPQFEKMTTSDPNPKPIVEDAQPEQITATECLAGLLENPTNLEGKLADAEEEIILEGEDTQASLNW SVIVPALVIVLATVVWGIGFKDSFTNFASSALSAVVDNLGWAFILFGTVFVFFIVVIAAS KFGTIRLGRIDEAPEFRTVSWISMMFAAGMGIGLMFYGTTEPLTFYRNGVPGHDEHNVGV AMSTTMFHWTLHPWAIYAIVGLAIAYSTFRVGRKQLLSSAFVPLIGEKGAEGWLGKLIDI LAIIATVFGTACSLGLGALQIGAGLSAANIIEDPSDWTIVGIVSVLTLAFIFSAISGVGK GIQYLSNANMVLAALLAIFVFVVGPTVSILNLLPGSIGNYLSNFFQMAGRTAMSADGTAG EWLGSWTIFYWAWWISWSPFVGMFLARISRGRSIREFILGVLLVPAGVSTVWFSIFGGTA IVFEQNGESIWGDGAAEEQLFGLLHALPGGQIMGIIAMILLGTFFITSADSASTVMGTMS QHGQLEANKWVTAAWGVATAAIGLTLLLSGGDNALSNLQNVTIVAATPFLFVVIGLMFAL VKDLSNDVIYLEYREQQRFNARLARERRVHNEHRKRELAAKRRDYKDDDK Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Molecular weight | Theoretical: 65.244039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: WSHPQFEKMT TSDPNPKPIV EDAQPEQITA TEELAGLLEN PTNLEGKLAD AEEEIILEGE DTQASLNWSV IVPALVIVLA TVVWGIGFK DSFTNFASSA LSAVVDNLGW AFILFGTVFV FFIVVIAASK FGTIRLGRID EAPEFRTVSW ISMMFAAGMG I GLMFYGTT ...String: WSHPQFEKMT TSDPNPKPIV EDAQPEQITA TEELAGLLEN PTNLEGKLAD AEEEIILEGE DTQASLNWSV IVPALVIVLA TVVWGIGFK DSFTNFASSA LSAVVDNLGW AFILFGTVFV FFIVVIAASK FGTIRLGRID EAPEFRTVSW ISMMFAAGMG I GLMFYGTT EPLTFYRNGV PGHDEHNVGV AMSTTMFHWT LHPWAIYAIV GLAIAYSTFR VGRKQLLSSA FVPLIGEKGA EG WLGKLID ILAIIATVFG TACSLGLGAL QIGAGLSAAN IIEDPSDWTI VGIVSVLTLA FIFSAISGVG KGIQYLSNAN MVL AALLAI FVFVVGPTVS ILNLLPGSIG NYLSNFFQMA GRTAMSADGT AGEWLGSWTI FYWAWWISWS PFVGMFLARI SRGR SIREF ILGVLLVPAG VSTVWFSIFG GTAIVFEQNG ESIWGDGAAE EQLFGLLHAL PGGQIMGIIA MILLGTFFIT SADSA STVM GTMSQHGQLE ANKWVTAAWG VATAAIGLTL LLSGGDNALS NLQNVTIVAA TPFLFVVIGL MFALVKDLSN DVIYLE YRE QQRFNARLAR ERRVHNEHRK RELAAKRRCE RKASGAGKRR UniProtKB: Glycine betaine transporter BetP |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 3 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: TRIMETHYL GLYCINE
| Macromolecule | Name: TRIMETHYL GLYCINE / type: ligand / ID: 3 / Number of copies: 3 / Formula: BET |
|---|---|
| Molecular weight | Theoretical: 118.154 Da |
| Chemical component information |  ChemComp-BET: |
-Macromolecule #4: dodecyl beta-D-glucopyranoside
| Macromolecule | Name: dodecyl beta-D-glucopyranoside / type: ligand / ID: 4 / Number of copies: 2 / Formula: XKJ |
|---|---|
| Molecular weight | Theoretical: 348.475 Da |
| Chemical component information | 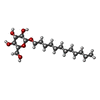 ChemComp-XKJ: |
-Macromolecule #5: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 5 / Number of copies: 1 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #6: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 6 / Number of copies: 2 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Macromolecule #7: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 7 / Number of copies: 1 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 100 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 200 |
|---|---|
| Temperature | Min: 96.0 K |
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 3929 / Average exposure time: 5.88 sec. / Average electron dose: 53.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































