[English] 日本語
 Yorodumi
Yorodumi- EMDB-50339: Cryo-EM structure of Trypanosoma cruzi glycosomal malate dehydrogenase -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Trypanosoma cruzi glycosomal malate dehydrogenase | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Oxidoreductase / Tetramer / Metabolic enzyme | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationintracellular organelle lumen / (S)-malate dehydrogenase (NAD+, oxaloacetate-forming) / L-malate dehydrogenase (NAD+) activity / carboxylic acid metabolic process / tricarboxylic acid cycle / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | ||||||||||||
 Authors Authors | Lipinski O / Sonani RR / Blat A / Jemiola-Rzeminska M / Patel SN / Sood T / Dubin G | ||||||||||||
| Funding support |  Poland, 3 items Poland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structure of Trypanosoma peroxisomal import complex unveils conformational heterogeneity. Authors: Ravi R Sonani / Artur Blat / Malgorzata Jemiola-Rzeminska / Oskar Lipinski / Stuti N Patel / Tabassum Sood / Grzegorz Dubin /    Abstract: Peroxisomes are membrane enclosed organelles hosting diverse metabolic processes in eukaryotic cells. Having no protein synthetic abilities, peroxisomes import all required enzymes from the cytosol ...Peroxisomes are membrane enclosed organelles hosting diverse metabolic processes in eukaryotic cells. Having no protein synthetic abilities, peroxisomes import all required enzymes from the cytosol through a peroxin (Pex) import system. Peroxisome targeting sequence 1 (PTS1)-tagged cargo is recognized by cytosolic receptor, Pex5. The cargo-Pex5 complex docks at the peroxisomal membrane translocon, composed of Pex14 and Pex13, facilitating translocation into the peroxisomal lumen. Despite its significance, the structural basis of the process is only partially understood. Here, we characterize the cargo-Pex5-Pex14 ternary complex from Trypanosoma cruzi. Cryo-electron microscopy maps enabled model building for Pex5 (residues 327-462 and 487-653) bound to malate dehydrogenase (MDH; residues 1-323) cargo tetramer and Pex14 (residues 21-85). The model provides insight into conformational heterogeneity and identifies secondary interfaces. Specifically, we observe that orientations of Pex5 relative to MDH span a 17° angle. Additionally, PTS1- and Wxxx(F/Y)-independent contact surfaces are observed at MDH-Pex5 and Pex5-Pex14 interfaces, respectively. Mutational analysis indicates that the non-PTS1 MDH-Pex5 interface does not significantly contribute to the affinity, but limits the conformational heterogeneity of MDH-Pex5 interface. The Pex5-Pex14 interface constitutes an extended binding site of Pex14 over Pex5. We discuss the implications of these findings for understanding peroxisomal import mechanism. #1:  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Structure of Trypanosoma peroxisomal import complex unveils conformational dynamics Authors: Sonani RR / Blat A / Jemiola-Rzeminska M / Lipinski O / Patel SN / Sood T / Dubin G | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50339.map.gz emd_50339.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50339-v30.xml emd-50339-v30.xml emd-50339.xml emd-50339.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50339.png emd_50339.png | 172.7 KB | ||
| Filedesc metadata |  emd-50339.cif.gz emd-50339.cif.gz | 6.2 KB | ||
| Others |  emd_50339_half_map_1.map.gz emd_50339_half_map_1.map.gz emd_50339_half_map_2.map.gz emd_50339_half_map_2.map.gz | 37.8 MB 37.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50339 http://ftp.pdbj.org/pub/emdb/structures/EMD-50339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50339 | HTTPS FTP |
-Validation report
| Summary document |  emd_50339_validation.pdf.gz emd_50339_validation.pdf.gz | 1002.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50339_full_validation.pdf.gz emd_50339_full_validation.pdf.gz | 1002.4 KB | Display | |
| Data in XML |  emd_50339_validation.xml.gz emd_50339_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_50339_validation.cif.gz emd_50339_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50339 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50339 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50339 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50339 | HTTPS FTP |
-Related structure data
| Related structure data |  9feeMC  9fefC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50339.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50339.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
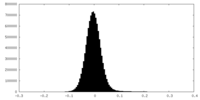
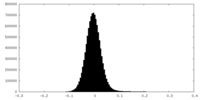
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_50339_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
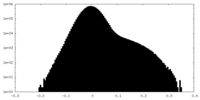
| Density Histograms |
-Half map: #2
| File | emd_50339_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
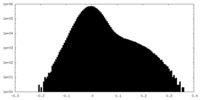
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric complex of glycosomal malate dehydrogenase
| Entire | Name: Tetrameric complex of glycosomal malate dehydrogenase |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric complex of glycosomal malate dehydrogenase
| Supramolecule | Name: Tetrameric complex of glycosomal malate dehydrogenase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: malate dehydrogenase
| Macromolecule | Name: malate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO EC number: (S)-malate dehydrogenase (NAD+, oxaloacetate-forming) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.13798 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHHHHHHMV NVAVIGAAGG IGQSLSLLLL RELPFGSTLS LYDVVGAPGV AADLSHIDRA GITVKHAAGK LPPVPRDPAL TELAEGVDV FVIVAGVPRK PGMTRDDLFN VNAGIVMDLV LTCASVSPNA CFCIVTNPVN STTPIAAQTL RKIGVYNKNK L LGVSLLDG ...String: MAHHHHHHMV NVAVIGAAGG IGQSLSLLLL RELPFGSTLS LYDVVGAPGV AADLSHIDRA GITVKHAAGK LPPVPRDPAL TELAEGVDV FVIVAGVPRK PGMTRDDLFN VNAGIVMDLV LTCASVSPNA CFCIVTNPVN STTPIAAQTL RKIGVYNKNK L LGVSLLDG LRATRFINNA RHPLVVPYVP VVGGHSDVTI VPLYSQIPGP LPDESTLKEI RKRVQVAGTE VVKAKAGRGS AT LSMAEAG ARFTMHVVKA LMGLDTPMVY AYVDTDGEHE CPFLAMPVVL GKNGIERRLP IGPITTVEKE MLEEAVGVVK KNI AKGETF ARSKL UniProtKB: malate dehydrogenase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 42.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)