[English] 日本語
 Yorodumi
Yorodumi- EMDB-49745: Cryo-EM structure of Gi-coupled FFA2 in complex with TUG-1375 and... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Gi-coupled FFA2 in complex with TUG-1375 and compound 187 | |||||||||
 Map data Map data | FFA2-Gi_TUG1375-cmpd187 map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / agonist / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of acute inflammatory response to non-antigenic stimulus / regulation of peptide hormone secretion / regulation of acute inflammatory response / mucosal immune response / leukocyte chemotaxis involved in inflammatory response / Free fatty acid receptors / cell surface pattern recognition receptor signaling pathway / positive regulation of cytokine production involved in immune response / lipid storage / cellular response to fatty acid ...positive regulation of acute inflammatory response to non-antigenic stimulus / regulation of peptide hormone secretion / regulation of acute inflammatory response / mucosal immune response / leukocyte chemotaxis involved in inflammatory response / Free fatty acid receptors / cell surface pattern recognition receptor signaling pathway / positive regulation of cytokine production involved in immune response / lipid storage / cellular response to fatty acid / fat cell differentiation / ligand-gated ion channel signaling pathway / adenylate cyclase inhibitor activity / positive regulation of chemokine production / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / positive regulation of interleukin-8 production / cell projection / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / positive regulation of insulin secretion / response to peptide hormone / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / glucose homeostasis / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / synapse / centrosome Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang X / Tikhonova I / Milligan G / Zhang C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: Allosteric modulation and biased signalling at free fatty acid receptor 2. Authors: Xuan Zhang / Abdul-Akim Guseinov / Laura Jenkins / Alice Valentini / Sara Marsango / Katrine Schultz-Knudsen / Trond Ulven / Elisabeth Rexen Ulven / Irina G Tikhonova / Graeme Milligan / Cheng Zhang /    Abstract: Free fatty acid receptor 2 (FFA2) is a G protein-coupled receptor (GPCR) that is a primary sensor for short-chain fatty acids produced by gut microbiota. Consequently, FFA2 is a promising drug ...Free fatty acid receptor 2 (FFA2) is a G protein-coupled receptor (GPCR) that is a primary sensor for short-chain fatty acids produced by gut microbiota. Consequently, FFA2 is a promising drug target for immunometabolic disorders. Here we report cryogenic electronic microscopy structures of FFA2 in complex with two G proteins and three distinct classes of positive allosteric modulators (PAMs), and describe noncanonical activation mechanisms that involve conserved structural features of class A GPCRs. Two PAMs disrupt the E/DRY activation microswitch and stabilize the conformation of intracellular loop 2 by binding to lipid-facing pockets near the cytoplasmic side of the receptor. By contrast, the third PAM promotes the separation of transmembrane helices 6 and 7 by interacting with transmembrane helix 6 at the receptor-lipid interface. Molecular dynamic simulations and mutagenesis experiments confirm these noncanonical activation mechanisms. Furthermore, we demonstrate the molecular basis for the G versus G bias, which is due to distinct conformations of intracellular loop 2 stabilized by different PAMs. These findings provide a framework for the design of tailored GPCR modulators, with implications that extend beyond FFA2 to the broader field of GPCR drug discovery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_49745.map.gz emd_49745.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-49745-v30.xml emd-49745-v30.xml emd-49745.xml emd-49745.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_49745.png emd_49745.png | 87.9 KB | ||
| Filedesc metadata |  emd-49745.cif.gz emd-49745.cif.gz | 7.3 KB | ||
| Others |  emd_49745_half_map_1.map.gz emd_49745_half_map_1.map.gz emd_49745_half_map_2.map.gz emd_49745_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-49745 http://ftp.pdbj.org/pub/emdb/structures/EMD-49745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49745 | HTTPS FTP |
-Related structure data
| Related structure data |  9ns9MC  9clwC  9cm3C  9cm7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_49745.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_49745.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FFA2-Gi_TUG1375-cmpd187 map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.72 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: FFA2-Gi TUG1375-cmpd187 map half A
| File | emd_49745_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FFA2-Gi_TUG1375-cmpd187_map_half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: FFA2-Gi TUG1375-cmpd187 map half B
| File | emd_49745_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FFA2-Gi_TUG1375-cmpd187_map_half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TUG-1375-compound 187-FFA2-Gi complex
| Entire | Name: TUG-1375-compound 187-FFA2-Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: TUG-1375-compound 187-FFA2-Gi complex
| Supramolecule | Name: TUG-1375-compound 187-FFA2-Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Free fatty acid receptor 2
| Macromolecule | Name: Free fatty acid receptor 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.023004 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDGQP GNGSAFLLAP NGSHAPDHNV TQQRDEENLY FQGVDMLPDW KSSLILMAYI IIFLTGLPAN LLALRAFVGR IRQPQPAPV HILLLSLTLA DLLLLLLLPF KIIEAASNFR WYLPKVVCAL TSFGFYSSIY CSTWLLAGIS IERYLGVAFP V QYKLSRRP ...String: DYKDDDDGQP GNGSAFLLAP NGSHAPDHNV TQQRDEENLY FQGVDMLPDW KSSLILMAYI IIFLTGLPAN LLALRAFVGR IRQPQPAPV HILLLSLTLA DLLLLLLLPF KIIEAASNFR WYLPKVVCAL TSFGFYSSIY CSTWLLAGIS IERYLGVAFP V QYKLSRRP LYGVIAALVA WVMSFGHCTI VIIVQYLNTT EQVRSGNEIT CYENFTDNQL DVVLPVRLEL CLVLFFIPMA VT IFCYWRF VWIMLSQPLV GAQRRRRAVG LAVVTLLNFL VCFGPYNVSH LVGYHQRKSP WWRSIAVVFS SLNASLDPLL FYF SSSVVR RAFGRGLQVL RNQGSSLLGR RGKDTAEGTN EDRGVGQGEG MPSSDFTTEA AAVFTLEDFV GDWEQTAAYN LDQV LEQGG VSSLLQNLAV SVTPIQRIVR SGENALKIDI HVIIPYEGLS ADQMAQIEEV FKVVYPVDDH HFKVILPYGT LVIDG VTPN MLNYFGRPYE GIAVFDGKKI TVTGTLWNGN KIIDERLITP DGSMLFRVTI NSHHHHHHHH UniProtKB: Free fatty acid receptor 2 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.845559 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWNGSSGGG GSGGGGSSGV SGWRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.634797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL LEENLYFQGA SHHHHHHHH |
-Macromolecule #6: (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phe...
| Macromolecule | Name: (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: 9UJ |
|---|---|
| Molecular weight | Theoretical: 442.915 Da |
| Chemical component information | 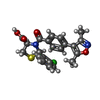 ChemComp-9UJ: |
-Macromolecule #7: 4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-7-(2-fluorobenzene-1-sulfo...
| Macromolecule | Name: 4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-7-(2-fluorobenzene-1-sulfonyl)-2-methyl-5H-pyrrolo[3,2-d]pyrimidin-6-amine type: ligand / ID: 7 / Number of copies: 1 / Formula: A1AZC |
|---|---|
| Molecular weight | Theoretical: 419.473 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































