+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MNV Allosteric escape mutant V339I + GCDCA | |||||||||
 Map data Map data | MNV1 Allosteric Escape mutant V339I GCDCA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | norovirus / escape / allosteric / mutant / VIRUS | |||||||||
| Function / homology | Calicivirus coat protein C-terminal / Calicivirus coat protein C-terminal / Calicivirus coat protein / Calicivirus coat protein / virion component / Picornavirus/Calicivirus coat protein / Viral coat protein subunit / host cell cytoplasm / Capsid protein VP1 Function and homology information Function and homology information | |||||||||
| Biological species |   Murine norovirus 1 Murine norovirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Smith TJ / Sherman M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2025 Journal: J Virol / Year: 2025Title: Murine norovirus allosteric escape mutants mimic gut activation. Authors: Michael B Sherman / Hong Q Smith / Faith Cox / Christiane E Wobus / Gillian C Lynch / B Montgomery Pettitt / Thomas J Smith /  Abstract: Murine norovirus (MNV) undergoes large conformational changes in response to the environment. The T=3 icosahedral capsid is composed of 180 copies of ~58 kDa VP1 that has N-terminal (N), shell (S), ...Murine norovirus (MNV) undergoes large conformational changes in response to the environment. The T=3 icosahedral capsid is composed of 180 copies of ~58 kDa VP1 that has N-terminal (N), shell (S), and C-terminal protruding (P) domains. In phosphate-buffered saline, the P domains are loosely tethered to the shell and float ~15 Å above the surface. At conditions found in the gut (i.e., low pH with high metal ion and bile salt concentrations), the P domain rotates and drops onto the shell with intra P domain changes that enhance receptor interactions while blocking antibody binding. Two of our monoclonal antibodies (2D3 and 4F9) have broad strain recognition, and the only escape mutants, V339I and D348E, are located on the C'D' loop and ~20 Å from the epitope. Here, we determined the cryo-EM structures of V339I and D348E at neutral pH +/-metal ions and bile salts. These allosteric escape mutants have the activated conformation in the absence of gut triggers. Since this conformation is not recognized by antibodies, it explains how these mutants evade antibody recognition. Dynamic simulations of the P domain further suggest that movement of the C'D' loop may be the rate-limiting step in the conformational change and that V339I increases the motion of the A'B'/E'F' loops compared to the wild-type (WT), facilitating the transition to the activated state. These findings have important implications for norovirus vaccine design since they uncover a form of the viral capsid that should lend superior immune protection against subsequent challenge by wild-type virus.IMPORTANCEImmune protection from norovirus infection is notoriously transient in both humans and mice. Our results strongly suggest that this is likely because the "activated" form of the virus found in gut conditions is not recognized by antibodies created in the circulation. By reversibly presenting one structure in the gut and a completely different antigenic structure in circulation, the gut tissue can be infected in subsequent challenges, while extraintestinal organs are protected. We find here that allosteric escape mutants to the most broadly neutralizing antibodies thwart recognition by transitioning to the activated state without the need for gut triggers (i.e., bile, low pH, or metal ions). These findings are significant because it is now feasible to present the activated form of the virus to the immune system (for example, as a vaccine) to better protect the gut tissue for longer periods of time. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47839.map.gz emd_47839.map.gz | 567.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47839-v30.xml emd-47839-v30.xml emd-47839.xml emd-47839.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_47839.png emd_47839.png | 247.6 KB | ||
| Filedesc metadata |  emd-47839.cif.gz emd-47839.cif.gz | 5.7 KB | ||
| Others |  emd_47839_half_map_1.map.gz emd_47839_half_map_1.map.gz emd_47839_half_map_2.map.gz emd_47839_half_map_2.map.gz | 555.3 MB 555.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47839 http://ftp.pdbj.org/pub/emdb/structures/EMD-47839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47839 | HTTPS FTP |
-Validation report
| Summary document |  emd_47839_validation.pdf.gz emd_47839_validation.pdf.gz | 1015 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_47839_full_validation.pdf.gz emd_47839_full_validation.pdf.gz | 1014.6 KB | Display | |
| Data in XML |  emd_47839_validation.xml.gz emd_47839_validation.xml.gz | 19.6 KB | Display | |
| Data in CIF |  emd_47839_validation.cif.gz emd_47839_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47839 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47839 | HTTPS FTP |
-Related structure data
| Related structure data |  9eapMC  9eanC  9eaoC  9eaqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47839.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47839.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MNV1 Allosteric Escape mutant V339I GCDCA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: MNV1 Allosteric Escape mutant V339I GCDCA
| File | emd_47839_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MNV1 Allosteric Escape mutant V339I GCDCA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: MNV1 Allosteric Escape mutant V339I GCDCA
| File | emd_47839_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MNV1 Allosteric Escape mutant V339I GCDCA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Murine norovirus 1
| Entire | Name:   Murine norovirus 1 Murine norovirus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Murine norovirus 1
| Supramolecule | Name: Murine norovirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 223997 / Sci species name: Murine norovirus 1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Murine norovirus 1 Murine norovirus 1 |
| Molecular weight | Theoretical: 58.583434 KDa |
| Sequence | String: RMSDGAAPKA NGSEASGQDL VPAAVEQAVP IQPVAGAALA APAAGQINQI DPWIFQNFVQ CPLGEFSISP RNTPGEILFD LALGPGLNP YLAHLSAMYT GWVGNMEVQL VLAGNAFTAG KVVVALVPPY FPKGSLTTAQ ITCFPHVMCD VRTLEPIQLP L LDVRRVLW ...String: RMSDGAAPKA NGSEASGQDL VPAAVEQAVP IQPVAGAALA APAAGQINQI DPWIFQNFVQ CPLGEFSISP RNTPGEILFD LALGPGLNP YLAHLSAMYT GWVGNMEVQL VLAGNAFTAG KVVVALVPPY FPKGSLTTAQ ITCFPHVMCD VRTLEPIQLP L LDVRRVLW HATQDQEESM RLVCMLYTPL RTNSPGDESF VVSGRLLSKP AADFNFVYLT PPIERTIYRM VDLPVIQPRL CT HARWPAP VYGLLVDPSL PSNPQWQNGR VHVDGTLLGT TPISGSWVSC FAAEAAYEFQ SGTGEVATFT LIEQDGSAYV PGD RAAPLG YPDFSGQLEI EIQTETTKTG DKLKVTTFEM ILGPTTNADQ APYQGRVFAS VTAAASLDLV DGRVRAVPRS IYGF QDTIP EYNDGLLVPL APPIGPFLPG EVLLRFRTYM RQIDTADAAA EAIDCALPQE FVSWFASNAF TVQSEALLLR YRNTL TGQL LFECKLYNEG YIALSYSGSG PLTFPTDGIF EVVSWVPRLY QLASVGSLAT GRMLKQ UniProtKB: Capsid protein VP1 |
-Macromolecule #2: GLYCOCHENODEOXYCHOLIC ACID
| Macromolecule | Name: GLYCOCHENODEOXYCHOLIC ACID / type: ligand / ID: 2 / Number of copies: 3 / Formula: CHO |
|---|---|
| Molecular weight | Theoretical: 449.623 Da |
| Chemical component information | 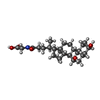 ChemComp-CHO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)