[English] 日本語
 Yorodumi
Yorodumi- EMDB-47293: Cryo-EM structure of human OATP1C1 F240A mutant in complex with e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human OATP1C1 F240A mutant in complex with estrone 3-sulfate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC transporter / E1S / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of thyroid hormone generation / Organic anion transport by SLCO transporters / sodium-independent organic anion transport / : / thyroid hormone transmembrane transporter activity / thyroid hormone transport / : / bile acid transmembrane transporter activity / bile acid and bile salt transport / transport across blood-brain barrier ...positive regulation of thyroid hormone generation / Organic anion transport by SLCO transporters / sodium-independent organic anion transport / : / thyroid hormone transmembrane transporter activity / thyroid hormone transport / : / bile acid transmembrane transporter activity / bile acid and bile salt transport / transport across blood-brain barrier / monoatomic ion transport / transmembrane transport / basolateral plasma membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Ge Y / Yadav GP / Jiang J / Huang P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2025 Journal: Cell / Year: 2025Title: Structural insights into brain thyroid hormone transport via MCT8 and OATP1C1. Authors: Yunhui Ge / Tongyi Dou / Thu Uyen Nguyen / Gaya P Yadav / Theodore G Wensel / Jiansen Jiang / Pengxiang Huang /  Abstract: Adequate delivery of thyroid hormones to the brain is crucial for normal neurological development. MCT8 and OATP1C1, two solute carrier (SLC) transporters, mediate the passage of thyroid hormones ...Adequate delivery of thyroid hormones to the brain is crucial for normal neurological development. MCT8 and OATP1C1, two solute carrier (SLC) transporters, mediate the passage of thyroid hormones across the blood-brain barrier and into the central nervous system. Mutations in MCT8 result in Allan-Herndon-Dudley syndrome (AHDS), an X-linked birth defect characterized by neurodevelopmental impairments and peripheral hyperthyroidism, whereas OATP1C1 deficiency is linked to brain hypometabolism and progressive neurodegeneration. Here, we report cryoelectron microscopy (cryo-EM) structures of MCT8 and OATP1C1 bound with the active thyroid hormone triiodothyronine (T3) and the prohormone thyroxine (T4) at 2.9 and 2.3 Å resolutions, respectively. Combined with functional studies, we elucidate their distinct thyroid hormone recognition and transport mechanisms and explain disease mutations. Although extracellular allosteric sites are not a common feature of SLC transporters, we identify one in OATP1C1. Collectively, these findings illuminate key aspects of thyroid hormone transport, a fundamental process in development and disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47293.map.gz emd_47293.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47293-v30.xml emd-47293-v30.xml emd-47293.xml emd-47293.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_47293_fsc.xml emd_47293_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_47293.png emd_47293.png | 54.9 KB | ||
| Masks |  emd_47293_msk_1.map emd_47293_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-47293.cif.gz emd-47293.cif.gz | 6.1 KB | ||
| Others |  emd_47293_half_map_1.map.gz emd_47293_half_map_1.map.gz emd_47293_half_map_2.map.gz emd_47293_half_map_2.map.gz | 98.3 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47293 http://ftp.pdbj.org/pub/emdb/structures/EMD-47293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47293 | HTTPS FTP |
-Related structure data
| Related structure data |  9dxoMC  9dwyC  9dxpC  9mr5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47293.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47293.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_47293_msk_1.map emd_47293_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_47293_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_47293_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human OATP1C1 F240A mutant in complex with estrone 3-sulfate
| Entire | Name: human OATP1C1 F240A mutant in complex with estrone 3-sulfate |
|---|---|
| Components |
|
-Supramolecule #1: human OATP1C1 F240A mutant in complex with estrone 3-sulfate
| Supramolecule | Name: human OATP1C1 F240A mutant in complex with estrone 3-sulfate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier organic anion transporter family member 1C1
| Macromolecule | Name: Solute carrier organic anion transporter family member 1C1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.463227 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDTSSKENIQ LFCKTSVQPV GRPSFKTEYP SSEEKQPCCG ELKVFLCALS FVYFAKALAE GYLKSTITQI ERRFDIPSSL VGVIDGSFE IGNLLVITFV SYFGAKLHRP KIIGAGCVIM GVGTLLIAMP QFFMEQYKYE RYSPSSNSTL SISPCLLESS S QLPVSVME ...String: MDTSSKENIQ LFCKTSVQPV GRPSFKTEYP SSEEKQPCCG ELKVFLCALS FVYFAKALAE GYLKSTITQI ERRFDIPSSL VGVIDGSFE IGNLLVITFV SYFGAKLHRP KIIGAGCVIM GVGTLLIAMP QFFMEQYKYE RYSPSSNSTL SISPCLLESS S QLPVSVME KSKSKISNEC EVDTSSSMWI YVFLGNLLRG IGETPIQPLG IAYLDDFASE DNAAFYIGCV QTVAIIGPIF GA LLGSLCA KLYVDIGFVN LDHITITPKD PQWVGAWWLG YLIAGIISLL AAVPFWYLPK SLPRSQSRED SNSSSEKSKF IID DHTDYQ TPQGENAKIM EMARDFLPSL KNLFGNPVYF LYLCTSTVQF NSLFGMVTYK PKYIEQQYGQ SSSRANFVIG LINI PAVAL GIFSGGIVMK KFRISVCGAA KLYLGSSVFG YLLFLSLFAL GCENSDVAGL TVSYQGTKPV SYHERALFSD CNSRC KCSE TKWEPMCGEN GITYVSACLA GCQTSNRSGK NIIFYNCTCV GIAASKSGNS SGIVGRCQKD NGCPQMFLYF LVISVI TSY TLSLGGIPGY ILLLRCIKPQ LKSFALGIYT LAIRVLAGIP APVYFGVLID TSCLKWGFKR CGSRGSCRLY DSNVFRH IY LGLTVILGTV SILLSIAVLF ILKKNYVSKH RSFITKRERT MVSTRFQKEN YTTSDHLLQP NYWPGKETQL AAALEVLF Q GPGAAEDQVD PRLIDGKHHH HHHHH UniProtKB: Solute carrier organic anion transporter family member 1C1 |
-Macromolecule #2: estrone 3-sulfate
| Macromolecule | Name: estrone 3-sulfate / type: ligand / ID: 2 / Number of copies: 3 / Formula: FY5 |
|---|---|
| Molecular weight | Theoretical: 350.429 Da |
| Chemical component information | 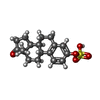 ChemComp-FY5: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Average electron dose: 50.83 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)