+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CCR6 bound by SQA1 and OXM2 | |||||||||
 Map data Map data | Cryo-EM structure of SQA1 and OXM2 bound to CCR6 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Antagonist / CCR6 / BRIL / MEMBRANE PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
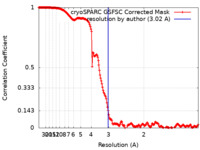
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Wasilko DJ / Wu H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for CCR6 modulation by allosteric antagonists. Authors: David Jonathan Wasilko / Brian S Gerstenberger / Kathleen A Farley / Wei Li / Jennifer Alley / Mark E Schnute / Ray J Unwalla / Jorge Victorino / Kimberly K Crouse / Ru Ding / Parag V ...Authors: David Jonathan Wasilko / Brian S Gerstenberger / Kathleen A Farley / Wei Li / Jennifer Alley / Mark E Schnute / Ray J Unwalla / Jorge Victorino / Kimberly K Crouse / Ru Ding / Parag V Sahasrabudhe / Fabien Vincent / Richard K Frisbie / Alpay Dermenci / Andrew Flick / Chulho Choi / Gary Chinigo / James J Mousseau / John I Trujillo / Philippe Nuhant / Prolay Mondal / Vincent Lombardo / Daniel Lamb / Barbara J Hogan / Gurdeep Singh Minhas / Elena Segala / Christine Oswald / Ian W Windsor / Seungil Han / Mathieu Rappas / Robert M Cooke / Matthew F Calabrese / Gabriel Berstein / Atli Thorarensen / Huixian Wu /   Abstract: The CC chemokine receptor 6 (CCR6) is a potential target for chronic inflammatory diseases. Previously, we reported an active CCR6 structure in complex with its cognate chemokine CCL20, revealing the ...The CC chemokine receptor 6 (CCR6) is a potential target for chronic inflammatory diseases. Previously, we reported an active CCR6 structure in complex with its cognate chemokine CCL20, revealing the molecular basis of CCR6 activation. Here, we present two inactive CCR6 structures in ternary complexes with different allosteric antagonists, CCR6/SQA1/OXM1 and CCR6/SQA1/OXM2. The oxomorpholine analogues, OXM1 and OXM2 are highly selective CCR6 antagonists which bind to an extracellular pocket and disrupt the receptor activation network. An energetically favoured U-shaped conformation in solution that resembles the bound form is observed for the active analogues. SQA1 is a squaramide derivative with close-in analogues reported as antagonists of chemokine receptors including CCR6. SQA1 binds to an intracellular pocket which overlaps with the G protein site, stabilizing a closed pocket that is a hallmark of inactive GPCRs. Minimal communication between the two allosteric pockets is observed, in contrast to the prevalent allosteric cooperativity model of GPCRs. This work highlights the versatility of GPCR antagonism by small molecules, complementing previous knowledge of CCR6 activation, and sheds light on drug discovery targeting CCR6. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_46533.map.gz emd_46533.map.gz | 506.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-46533-v30.xml emd-46533-v30.xml emd-46533.xml emd-46533.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_46533_fsc.xml emd_46533_fsc.xml | 17.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_46533.png emd_46533.png | 77.8 KB | ||
| Filedesc metadata |  emd-46533.cif.gz emd-46533.cif.gz | 7.2 KB | ||
| Others |  emd_46533_half_map_1.map.gz emd_46533_half_map_1.map.gz emd_46533_half_map_2.map.gz emd_46533_half_map_2.map.gz | 497.1 MB 497.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-46533 http://ftp.pdbj.org/pub/emdb/structures/EMD-46533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46533 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_46533.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_46533.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SQA1 and OXM2 bound to CCR6 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.654 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM half map 1
| File | emd_46533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EM half map 2
| File | emd_46533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CCR6-BRIL/Fab/Nb in complex with SQA1 and OXM2
| Entire | Name: CCR6-BRIL/Fab/Nb in complex with SQA1 and OXM2 |
|---|---|
| Components |
|
-Supramolecule #1: CCR6-BRIL/Fab/Nb in complex with SQA1 and OXM2
| Supramolecule | Name: CCR6-BRIL/Fab/Nb in complex with SQA1 and OXM2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: CCR6-BRIL/Fab/Nb complex co-purified with SQA1 and OXM2. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 kDa/nm |
-Macromolecule #1: Human CCR6
| Macromolecule | Name: Human CCR6 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.62302 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHE NLYFQGGTSV DSEMLLCSLQ EVRQFSRAFV PIAYSLICVF GLLGNILVV ITFAFYKKAR SMTDVYLANM AIADILFALT LPFWAVSHAT GAWVFSNATC KLLKGIYAIN FNCGMWLLTC I AMDRYIAI ...String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHE NLYFQGGTSV DSEMLLCSLQ EVRQFSRAFV PIAYSLICVF GLLGNILVV ITFAFYKKAR SMTDVYLANM AIADILFALT LPFWAVSHAT GAWVFSNATC KLLKGIYAIN FNCGMWLLTC I AMDRYIAI VQATKSFRLR SATLPRSKII CLVVWGLSVI ISSSTFVFNQ KYNTQGSDVC EPKYQTVSEP IRWKLLMLGL EL LFGFFIP LMFMIFCYTA IVKTLRRQLA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLEDK SPD SPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLERAR STLQKEVKAI RVIIAVVLVF LACQ IPHNM VLLVTAANLG KMNRSCQSEK LIAYTKTVTE VLAFLHCCLN PVLYAFIGQK FRNYFLKILK DLWCVRRKYK SSGFS |
-Macromolecule #2: anti-BRIL Fab Heavy chain
| Macromolecule | Name: anti-BRIL Fab Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.832098 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHSE ISEVQLVESG GGLVQPGGSL RLSCAASGFN VVDFSLHWVR QAPGKGLEWV AYISSSSGST SYADSVKGR FTISADTSKN TAYLQMNSLR AEDTAVYYCA RWGYWPGEPW WKAFDYWGQG TLVTVSSAST KGPSVFPLAP S SKSTSGGT ...String: MGWSCIILFL VATATGVHSE ISEVQLVESG GGLVQPGGSL RLSCAASGFN VVDFSLHWVR QAPGKGLEWV AYISSSSGST SYADSVKGR FTISADTSKN TAYLQMNSLR AEDTAVYYCA RWGYWPGEPW WKAFDYWGQG TLVTVSSAST KGPSVFPLAP S SKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KV DKKVEPK SGGSENLYFQ GSHHHHHHHH HH |
-Macromolecule #3: anti-BRIL Fab Nanobody
| Macromolecule | Name: anti-BRIL Fab Nanobody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.755214 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH HHENLYFQGS QVQLQESGGG LVQPGGSLRL SCAASGRTIS RYAMSWFRQA PGKEREFVAV ARRSGDGAFY ADSVQGRFT VSRDDAKNTV YLQMNSLKPE DTAVYYCAID SDTFYSGSYD YWGQGTQVTV SS |
-Macromolecule #4: anti-BRIL Fab Light chain
| Macromolecule | Name: anti-BRIL Fab Light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.343348 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHSS DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGT DFTLTISSLQ PEDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN F YPREAKVQ ...String: MGWSCIILFL VATATGVHSS DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGT DFTLTISSLQ PEDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN F YPREAKVQ WKVDNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRG |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #6: 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl...
| Macromolecule | Name: 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide type: ligand / ID: 6 / Number of copies: 1 / Formula: EBX |
|---|---|
| Molecular weight | Theoretical: 426.466 Da |
| Chemical component information | 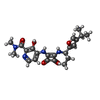 ChemComp-EBX: |
-Macromolecule #7: N-{[(2R)-4-(bicyclo[1.1.1]pentan-1-yl)-5-oxomorpholin-2-yl]methyl...
| Macromolecule | Name: N-{[(2R)-4-(bicyclo[1.1.1]pentan-1-yl)-5-oxomorpholin-2-yl]methyl}-1-[4-(trifluoromethyl)phenyl]cyclopropane-1-carboxamide type: ligand / ID: 7 / Number of copies: 1 / Formula: A1A1W |
|---|---|
| Molecular weight | Theoretical: 408.414 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM HEPES pH 7.5, 150 mM NaCl, 0.003% LMNG, 0.0003% CHS, 50 uM OXM2 and 50 uM of SQA1 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | CCR6-BRIL/Fab/Nb complex co-purified with SQA1 and OXM2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9d3e: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)