+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Best1 + GABA closed state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | calcium-activated chloride channel / GABA type A receptor / GABA-bound anion channel / channel-activator complex / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane microdomain / bicarbonate channel activity / transepithelial chloride transport / gamma-aminobutyric acid secretion, neurotransmission / detection of light stimulus involved in visual perception / ligand-gated channel activity / intracellularly calcium-gated chloride channel activity / glutamate secretion / bicarbonate transmembrane transporter activity / chloride transport ...membrane microdomain / bicarbonate channel activity / transepithelial chloride transport / gamma-aminobutyric acid secretion, neurotransmission / detection of light stimulus involved in visual perception / ligand-gated channel activity / intracellularly calcium-gated chloride channel activity / glutamate secretion / bicarbonate transmembrane transporter activity / chloride transport / chloride channel activity / chloride channel complex / regulation of calcium ion transport / visual perception / basal plasma membrane / protein complex oligomerization / regulation of synaptic plasticity / Stimuli-sensing channels / monoatomic ion transmembrane transport / presynapse / basolateral plasma membrane / identical protein binding / membrane / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||||||||
 Authors Authors | Owji AP / Kittredge A / Zhang Y / Yang T | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel. Authors: Jiali Wang / Aaron P Owji / Alec Kittredge / Zada Clark / Yu Zhang / Tingting Yang /  Abstract: Bestrophin-1 (Best1) is an anion channel genetically linked to vision-threatening retinal degenerative channelopathies. Here, we identify interactions between Best1 and both isoforms of glutamic acid ...Bestrophin-1 (Best1) is an anion channel genetically linked to vision-threatening retinal degenerative channelopathies. Here, we identify interactions between Best1 and both isoforms of glutamic acid decarboxylases (GAD65 and GAD67), elucidate the distinctive influences of GAD65 and GAD67 on Best1's permeability to various anions/neurotransmitters, discover the functionality of Best1 as a γ-Aminobutyric acid (GABA) type A receptor, and solve the structure of GABA-bound Best1. GAD65 and GAD67 both promote Best1-mediated Cl currents, but only GAD65 drastically enhances the permeability of Best1 to glutamate and GABA, for which GAD67 has no effect. GABA binds to Best1 on an extracellular site and stimulates Best1-mediated Cl currents at the nano-molar concentration level. The physiological role of GAD65 as a cell type-specific binding partner and facilitator of Best1 is demonstrated in retinal pigment epithelial cells. Together, our results reveal critical regulators of Best1 and inform a network of membrane transport metabolons formed between bestrophin channels and glutamate metabolic enzymes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45918.map.gz emd_45918.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45918-v30.xml emd-45918-v30.xml emd-45918.xml emd-45918.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_45918.png emd_45918.png | 110.6 KB | ||
| Masks |  emd_45918_msk_1.map emd_45918_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45918.cif.gz emd-45918.cif.gz | 6.4 KB | ||
| Others |  emd_45918_additional_1.map.gz emd_45918_additional_1.map.gz emd_45918_half_map_1.map.gz emd_45918_half_map_1.map.gz emd_45918_half_map_2.map.gz emd_45918_half_map_2.map.gz | 122.5 MB 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45918 http://ftp.pdbj.org/pub/emdb/structures/EMD-45918 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45918 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45918 | HTTPS FTP |
-Validation report
| Summary document |  emd_45918_validation.pdf.gz emd_45918_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45918_full_validation.pdf.gz emd_45918_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_45918_validation.xml.gz emd_45918_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_45918_validation.cif.gz emd_45918_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45918 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45918 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45918 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45918 | HTTPS FTP |
-Related structure data
| Related structure data |  9cttMC  9ctqC  9ctrC  9ctsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45918.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45918.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
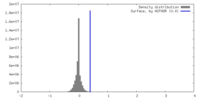
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45918_msk_1.map emd_45918_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
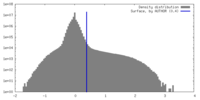
| Density Histograms |
-Additional map: full map
| File | emd_45918_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full_map | ||||||||||||
| Projections & Slices |
| ||||||||||||
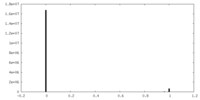
| Density Histograms |
-Half map: half A
| File | emd_45918_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
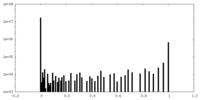
| Density Histograms |
-Half map: half B
| File | emd_45918_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GABA-bound Homopentameric complex of Best1 anion channel in fully...
| Entire | Name: GABA-bound Homopentameric complex of Best1 anion channel in fully open fffff state |
|---|---|
| Components |
|
-Supramolecule #1: GABA-bound Homopentameric complex of Best1 anion channel in fully...
| Supramolecule | Name: GABA-bound Homopentameric complex of Best1 anion channel in fully open fffff state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: GABA-activated structure |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 337.5 KDa |
-Macromolecule #1: Bestrophin-1
| Macromolecule | Name: Bestrophin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.629273 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TITYTSQVAN ARLGSFSRLL LCWRGSIYKL LYGEFLIFLL CYYIIRFIYR LALTEEQQLM FEKLTLYCDS YIQLIPISFV LGFYVTLVV TRWWNQYENL PWPDRLMSLV SGFVEGKDEQ GRLLRRTLIR YANLGNVLIL RSVSTAVYKR FPSAQHLVQA G FMTPAEHK ...String: TITYTSQVAN ARLGSFSRLL LCWRGSIYKL LYGEFLIFLL CYYIIRFIYR LALTEEQQLM FEKLTLYCDS YIQLIPISFV LGFYVTLVV TRWWNQYENL PWPDRLMSLV SGFVEGKDEQ GRLLRRTLIR YANLGNVLIL RSVSTAVYKR FPSAQHLVQA G FMTPAEHK QLEKLSLPHN MFWVPWVWFA NLSMKAWLGG RIRDPILLQS LLNEMNTLRT QCGHLYAYDW ISIPLVYTQV VT VAVYSFF LTCLVGRQFL NPAKAYPGHE LDLVVPVFTF LQFFFYVGWL KVAEQLINPF GEDDDDFETN WIVDRNLQVS LLA VDEMHQ DLPRMEPDMY WNKPEPQPPY TAASAQFRRA SFMGSTFNIS LNKEEMEFQP NQEDEEDAHA GIIGRFLGLQ SHDH HPPRA NSRTKLLWPK RESLLHEGLP KNHKAAKQNV RGQEDNKAWK LKAVDAFKSA PLYQRPGYYS APQTPLSPTP MFFPL EPSA PSKLHSVTGI DTKDKSLKTV SSGAKKSFEL LSESDGALME HPEVSQVRRK TVEFNLTDMP EIPENHLKEP LEQSPT NIH TTLKDHMDPY WALENRDEAH S UniProtKB: Bestrophin-1 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R0./1 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 713 / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: ab initio reconstruction |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 52529 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)