[English] 日本語
 Yorodumi
Yorodumi- EMDB-45308: CryoEM structure of Apo Cryptococcus neoformans H99 Acetyl-CoA Sy... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Apo Cryptococcus neoformans H99 Acetyl-CoA Synthetase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | trimer / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetate-CoA ligase / acetate-CoA ligase activity / acetyl-CoA biosynthetic process from acetate / AMP binding / ATP binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Cryptococcus neoformans var. grubii H99 (fungus) Cryptococcus neoformans var. grubii H99 (fungus) | |||||||||
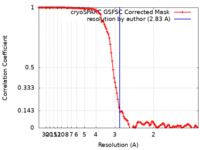
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Xu Z / Schnicker NJ / Jezeski AJ / Krysan DJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Not Published Journal: Not PublishedTitle: CryoEM structure of Apo Cryptococcus neoformans H99 Acetyl-CoA Synthetase Authors: Xu Z / Schnicker NJ / Jezeski AJ / Krysan DJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45308.map.gz emd_45308.map.gz | 136.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45308-v30.xml emd-45308-v30.xml emd-45308.xml emd-45308.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45308_fsc.xml emd_45308_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_45308.png emd_45308.png | 124.1 KB | ||
| Filedesc metadata |  emd-45308.cif.gz emd-45308.cif.gz | 6 KB | ||
| Others |  emd_45308_half_map_1.map.gz emd_45308_half_map_1.map.gz emd_45308_half_map_2.map.gz emd_45308_half_map_2.map.gz | 134.4 MB 134.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45308 http://ftp.pdbj.org/pub/emdb/structures/EMD-45308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45308 | HTTPS FTP |
-Related structure data
| Related structure data |  9c8rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45308.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45308.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
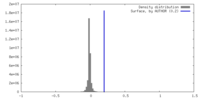
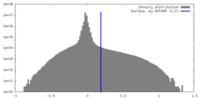
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_45308_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
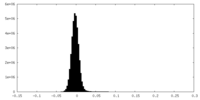
| Density Histograms |
-Half map: #2
| File | emd_45308_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
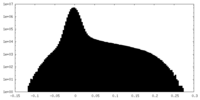
| Density Histograms |
- Sample components
Sample components
-Entire : Cryptococcus neoformans H99 Acetyl-CoA Synthetase
| Entire | Name: Cryptococcus neoformans H99 Acetyl-CoA Synthetase |
|---|---|
| Components |
|
-Supramolecule #1: Cryptococcus neoformans H99 Acetyl-CoA Synthetase
| Supramolecule | Name: Cryptococcus neoformans H99 Acetyl-CoA Synthetase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Cryptococcus neoformans var. grubii H99 (fungus) Cryptococcus neoformans var. grubii H99 (fungus) |
| Molecular weight | Theoretical: 238 KDa |
-Macromolecule #1: Acetyl-coenzyme A synthetase
| Macromolecule | Name: Acetyl-coenzyme A synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: acetate-CoA ligase |
|---|---|
| Source (natural) | Organism:  Cryptococcus neoformans var. grubii H99 (fungus) Cryptococcus neoformans var. grubii H99 (fungus) |
| Molecular weight | Theoretical: 77.47575 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHE NLYFQGKTEV APGVHHVHPL PDSVPESEDL FAPPPRMQGK EGRPKPHIGP NYESYVKEWA KTVGPNSDEW WAAKARETL DWYDDFKTVR AGGFEHGDVQ WFPEGTLNAA YNCLDRHYYK NPKKTAIIYE ADEPSESREV SYEELMQETC R VANVLKSY ...String: MHHHHHHHHE NLYFQGKTEV APGVHHVHPL PDSVPESEDL FAPPPRMQGK EGRPKPHIGP NYESYVKEWA KTVGPNSDEW WAAKARETL DWYDDFKTVR AGGFEHGDVQ WFPEGTLNAA YNCLDRHYYK NPKKTAIIYE ADEPSESREV SYEELMQETC R VANVLKSY GVKKGDAVSI YLPMTWQAAA AFLACARIGA IHSAVFAGFS AESLRDRVND CECKVLITTD EGRRGGKTIA TK QIVDAAL QQCPLVENVL VLRRTGNKVP MTEGRDKWWD EECAKMPAYC PCERMASEDP LFILYTSGST GKPKGVVHST AGY LLGTAL TLKYVFDAHP DDRFACMADI GWITGHSYII YGPLANGITT AVFESTPVYP TPSRYWDFVD KWKATQLYTA PTAI RLLRR MGEDHVKNHD LSSLRVLGSV GEPINPEAWH WYNDFAGKNQ CAIVDTYWMT ETGSISIAPL PGAISTKPGS ATFPF FGMD VDIIDPQTGQ VLEGNDVEGV LVARRPWPSI ARTVYRDHKR YLETYMKPYP GYFFFGDGAA RDYDGYMWIK GRVDDV INV SGHRLSTAEV ESALILHKGV AETAVVGCAD DLTGQAVYAF VTMKPEFDLK ATKEADLSKE LAIQVRKVIG PFAAPKK IY LVSDLPKTRS GKIMRRVLRK IVAGEGDQLG DLSSIADPQI VEEVKQKVTG SA UniProtKB: Acetyl-coenzyme A synthetase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 10 mM Tris pH7.5, 150 mM NaCl, 4mM MgCl2 and 10mM DTT | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)