+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Mnx H340A complex from Bacillus sp. PL-12 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / multicopper oxidase / manganese biomineralization / METAL TRANSPORT | |||||||||
| Function / homology | Multicopper oxidase, copper-binding site / Multicopper oxidases signature 2. / Multicopper oxidase, N-terminal / Multicopper oxidase / Cupredoxin / copper ion binding / MnxE / MnxF / MnxG Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
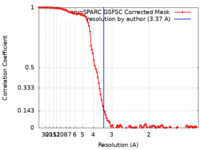
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Novikova IV / Evans JE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Am Chem Soc / Year: 2024 Journal: J Am Chem Soc / Year: 2024Title: Cryo-EM Structure of the Mnx Protein Complex Reveals a Tunnel Framework for the Mechanism of Manganese Biomineralization. Authors: Irina V Novikova / Alexandra V Soldatova / Trevor H Moser / Stephanie M Thibert / Christine A Romano / Mowei Zhou / Bradley M Tebo / James E Evans / Thomas G Spiro /  Abstract: The global manganese cycle relies on microbes to oxidize soluble Mn(II) to insoluble Mn(IV) oxides. Some microbes require peroxide or superoxide as oxidants, but others can use O directly, via ...The global manganese cycle relies on microbes to oxidize soluble Mn(II) to insoluble Mn(IV) oxides. Some microbes require peroxide or superoxide as oxidants, but others can use O directly, via multicopper oxidase (MCO) enzymes. One of these, MnxG from strain PL-12, was isolated in tight association with small accessory proteins, MnxE and MnxF. The protein complex, called Mnx, has eluded crystallization efforts, but we now report the 3D structure of a point mutant using cryo-EM single particle analysis, cross-linking mass spectrometry, and AlphaFold Multimer prediction. The β-sheet-rich complex features MnxG enzyme, capped by a heterohexameric ring of alternating MnxE and MnxF subunits, and a tunnel that runs through MnxG and its MnxEF cap. The tunnel dimensions and charges can accommodate the mechanistically inferred binuclear manganese intermediates. Comparison with the Fe(II)-oxidizing MCO, ceruloplasmin, identifies likely coordinating groups for the Mn(II) substrate, at the entrance to the tunnel. Thus, the 3D structure provides a rationale for the established manganese oxidase mechanism, and a platform for further experiments to elucidate mechanistic details of manganese biomineralization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45001.map.gz emd_45001.map.gz | 68.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45001-v30.xml emd-45001-v30.xml emd-45001.xml emd-45001.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45001_fsc.xml emd_45001_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_45001.png emd_45001.png | 113.4 KB | ||
| Masks |  emd_45001_msk_1.map emd_45001_msk_1.map | 137.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45001.cif.gz emd-45001.cif.gz | 6.4 KB | ||
| Others |  emd_45001_half_map_1.map.gz emd_45001_half_map_1.map.gz emd_45001_half_map_2.map.gz emd_45001_half_map_2.map.gz | 127.4 MB 127.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45001 http://ftp.pdbj.org/pub/emdb/structures/EMD-45001 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45001 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45001 | HTTPS FTP |
-Related structure data
| Related structure data |  9bxaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45001.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45001.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||
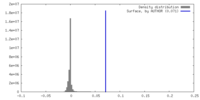
| Density |
| ||||||||||||||||||||||||||||||||||||
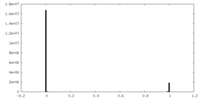
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45001_msk_1.map emd_45001_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
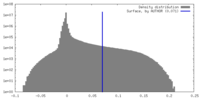
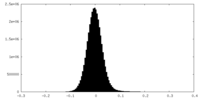
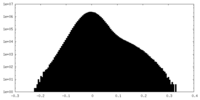
| Density Histograms |
-Half map: #2
| File | emd_45001_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
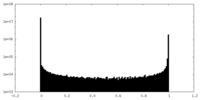
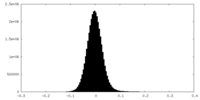
| Density Histograms |
-Half map: #1
| File | emd_45001_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
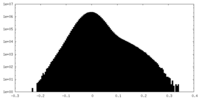
| Density Histograms |
- Sample components
Sample components
-Entire : Mnx H340A complex
| Entire | Name: Mnx H340A complex |
|---|---|
| Components |
|
-Supramolecule #1: Mnx H340A complex
| Supramolecule | Name: Mnx H340A complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: MnxG
| Macromolecule | Name: MnxG / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 138.340672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRKFHVVGI STRIVVNTFG DHNPNGRIYV LKENESKLKD LVRKNPYKPI DLVQPLAIRA NEGDIVEILF ENQLSFSAGM HFQEADYSV LSSDGADAGY NPDTTVEPGG EILYRLNVNQ EGICFFTDLG NVSSTEQGSS VQGLFGALLV QKRGSSWTDP V TGGPINSG ...String: MLRKFHVVGI STRIVVNTFG DHNPNGRIYV LKENESKLKD LVRKNPYKPI DLVQPLAIRA NEGDIVEILF ENQLSFSAGM HFQEADYSV LSSDGADAGY NPDTTVEPGG EILYRLNVNQ EGICFFTDLG NVSSTEQGSS VQGLFGALLV QKRGSSWTDP V TGGPINSG VYADIHHPFL PSFREYAWFF NDEMEIRDLT GERPLNPMTN QEAESFHGVN LRYEPMTNRK RLMEAGVVCP DC DSEEVHH DSWVFGDPAT PILRGYVGDP AVIRLIHGGV KETHVFHYHV HQWLGDSSNI NAEILDAQSI SPQTHYSIQP LYG LGSLHG AIGDSIIHCH LYPAFGIGMW GMNRVFDTLQ DGSQCYPNGV RIKALMPLPD RPEPPKPTPE KPGFPNFIPG KVGY KAPRP PLGIVGGREM TELERNAAIE NPRPGAVFVD PCLDQDPVVV EFNVSAIEMP VVYNKQGWHD PKARFYVMDE DLDDI LSGK KEPEPLVFHV PAGTCIRMNY TNRMPHILDG DAFQLVTRTY ENGFHIHFVK FDVLACDGGN VGWNYDSAVL PGQTIR YEW YAETELKAFF FHDHLFANSH QQHGVFGAGV IQPRFSKFLD SRTGDEVDHG TQISVEHPLI PDYRDQTLFV HDFALLF DK NGRPIQPPEY PGSEDDPGVF GVNFKCEPLK FRLGEDCDPA YSFSSYVHGD PVTPILRAYE GDPIRIRLLQ GAHEESHS F NIHGLRWKEE RPDLGSSMKA QQHIGISESF TFETEIPASG DYLWAFEDEE DVWLGTWGLI RAYKGRMEDL IVLTDREAL PEGSAETPKP TGKPPEKANP LASLPPGAYQ GSPVKKFEVV AFQTPIQYNS YGDHDPYGII FALKEDVEDI LTGKKNPVPL ILRANVGDL VEVTLTSELK KELFPFQDGI HPYPPVKEQS FYPPSLRISL HTSLLNYDVK TSSGDTVGYN PDQTVGPGET I TYRWFVDG QFGMCSMWDM ADLRNHRSFG TFGAFVAESR FTTYLDPYSL EKAITGENVI LRHPLLPATR EFVLILHDGV RL EDKDGKV IIDPMDGVVP DTEELEEVDT YDYGSRGFNY RSERLINRYK EHPVMHELFS SEVFGDPATP LFEAYPGEPV VMR ITTPAE RRRAHTFHLH GHYWKFDSKD LDSRIQSFLG HMVTGHTDDL RLIGGAGGVF NFPGDYLYRS GNIRWDIELG MWGI FRVHK DSKENLPRLE EVEGGWDNEE KA UniProtKB: MnxG |
-Macromolecule #2: MnxE
| Macromolecule | Name: MnxE / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.185235 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHDSPLKSLS AASNVASVND PLFDFFNKHM GKQILIITES SQLNILGQTF RPIFCGKVAE VEPGHLTLSP VTIKILNAPF HKFPIPLSI PFEKIAHFTT DVDCSMRIPL V UniProtKB: MnxE |
-Macromolecule #3: MnxF
| Macromolecule | Name: MnxF / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.026734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEALFPMSTD YSKMTDVNEI HDSAILEHFR NGIGHKTLVI SPSYPYMFVG IIKELIGDTV MIDVETTHFA QLENREWYIH IHNIEVFYI ERPGAPKIPK LEDY UniProtKB: MnxF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % |
| Details | contains 80mM CHAPSO |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)