[English] 日本語
 Yorodumi
Yorodumi- EMDB-44602: Cryo-EM structure of the mammalian peptide transporter PepT2 boun... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 2 | |||||||||||||||
 Map data Map data | unsharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | protein-coupled peptide transporter / peptide transport / antibiotics / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhigh-affinity oligopeptide transmembrane transporter activity / tripeptide import across plasma membrane / oligopeptide transport / dipeptide transport / peptidoglycan transport / dipeptide import across plasma membrane / tripeptide transmembrane transporter activity / metanephric proximal tubule development / peptide:proton symporter activity / antibacterial innate immune response ...high-affinity oligopeptide transmembrane transporter activity / tripeptide import across plasma membrane / oligopeptide transport / dipeptide transport / peptidoglycan transport / dipeptide import across plasma membrane / tripeptide transmembrane transporter activity / metanephric proximal tubule development / peptide:proton symporter activity / antibacterial innate immune response / dipeptide transmembrane transporter activity / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / xenobiotic transport / renal absorption / phagocytic vesicle membrane / protein transport / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
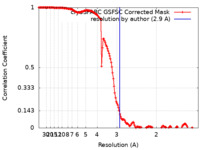
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||
 Authors Authors | Parker JL / Deme JC / Lea SM / Newstead S | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for antibiotic transport and inhibition in PepT2. Authors: Joanne L Parker / Justin C Deme / Simon M Lichtinger / Gabriel Kuteyi / Philip C Biggin / Susan M Lea / Simon Newstead /   Abstract: The uptake and elimination of beta-lactam antibiotics in the human body are facilitated by the proton-coupled peptide transporters PepT1 (SLC15A1) and PepT2 (SLC15A2). The mechanism by which SLC15 ...The uptake and elimination of beta-lactam antibiotics in the human body are facilitated by the proton-coupled peptide transporters PepT1 (SLC15A1) and PepT2 (SLC15A2). The mechanism by which SLC15 family transporters recognize and discriminate between different drug classes and dietary peptides remains unclear, hampering efforts to improve antibiotic pharmacokinetics through targeted drug design and delivery. Here, we present cryo-EM structures of the proton-coupled peptide transporter, PepT2 from Rattus norvegicus, in complex with the widely used beta-lactam antibiotics cefadroxil, amoxicillin and cloxacillin. Our structures, combined with pharmacophore mapping, molecular dynamics simulations and biochemical assays, establish the mechanism of beta-lactam antibiotic recognition and the important role of protonation in drug binding and transport. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44602.map.gz emd_44602.map.gz | 266.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44602-v30.xml emd-44602-v30.xml emd-44602.xml emd-44602.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44602_fsc.xml emd_44602_fsc.xml | 16.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_44602.png emd_44602.png | 120.4 KB | ||
| Masks |  emd_44602_msk_1.map emd_44602_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44602.cif.gz emd-44602.cif.gz | 7.3 KB | ||
| Others |  emd_44602_additional_2.map.gz emd_44602_additional_2.map.gz emd_44602_half_map_1.map.gz emd_44602_half_map_1.map.gz emd_44602_half_map_2.map.gz emd_44602_half_map_2.map.gz | 293.9 MB 301.2 MB 301.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44602 http://ftp.pdbj.org/pub/emdb/structures/EMD-44602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44602 | HTTPS FTP |
-Related structure data
| Related structure data |  9biuMC  9birC  9bisC  9bitC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44602.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44602.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.732 Å | ||||||||||||||||||||||||||||||||||||
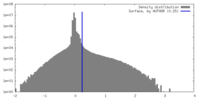
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_44602_msk_1.map emd_44602_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
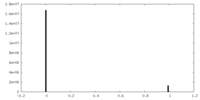
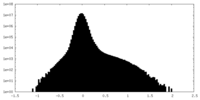
| Density Histograms |
-Additional map: unsharpened map
| File | emd_44602_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
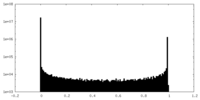
| Density Histograms |
-Half map: unsharpened map
| File | emd_44602_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
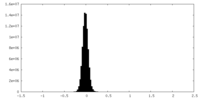
| Density Histograms |
-Half map: unsharpened map
| File | emd_44602_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the mammalian peptide transporter PepT2 with nanobody ...
| Entire | Name: Complex of the mammalian peptide transporter PepT2 with nanobody and bound cloxacillin, pose 2 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the mammalian peptide transporter PepT2 with nanobody ...
| Supramolecule | Name: Complex of the mammalian peptide transporter PepT2 with nanobody and bound cloxacillin, pose 2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: PepT2
| Supramolecule | Name: PepT2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: nanobody
| Supramolecule | Name: nanobody / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier family 15 member 2
| Macromolecule | Name: Solute carrier family 15 member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.477766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNPFQKNESK ETLFSPVSTE EMLPRPPSPP KKSPPKIFGS SYPVSIAFIV VNEFCERFSY YGMKAVLTLY FLYFLHWNED TSTSVYHAF SSLCYFTPIL GAAIADSWLG KFKTIIYLSL VYVLGHVFKS LGAIPILGGK MLHTILSLVG LSLIALGTGG I KPCVAAFG ...String: MNPFQKNESK ETLFSPVSTE EMLPRPPSPP KKSPPKIFGS SYPVSIAFIV VNEFCERFSY YGMKAVLTLY FLYFLHWNED TSTSVYHAF SSLCYFTPIL GAAIADSWLG KFKTIIYLSL VYVLGHVFKS LGAIPILGGK MLHTILSLVG LSLIALGTGG I KPCVAAFG GDQFEEEHAE ARTRYFSVFY LAINAGSLIS TFITPMLRGD VKCFGQDCYA LAFGVPGLLM VLALVVFAMG SK MYRKPPP EGNIVAQVIK CIWFALCNRF RNRSGDLPKR QHWLDWAAEK YPKHLIADVK ALTRVLFLYI PLPMFWALLD QQG SRWTLQ ANKMNGDLGF FVLQPDQMQV LNPFLVLIFI PLFDLVIYRL ISKCRINFSS LRKMAVGMIL ACLAFAVAAL VETK INGMI HPQPASQEIF LQVLNLADGD VKVTVLGSRN NSLLVESVSS FQNTTHYSKL HLEAKSQDLH FHLKYNSLSV HNDHS VEEK NCYQLLIHQD GESISSMLVK DTGIKPANGM AAIRFINTLH KDLNISLDTD APLSVGKDYG VSAYRTVLRG KYPAVH CET EDKVFSLDLG QLDFGTTYLF VITNITSQGL QAWKAEDIPV NKLSIAWQLP QYVLVTAAEV MFSVTGLEFS YSQAPSS MK SVLQAAWLLT VAVGNIIVLV VAQFSGLAQW AEFVLFSCLL LVVCLIFSVM AYYYVPLKSE DTREATDKQI PAVQGNMI N LETKNTRLVE GENLYFQ UniProtKB: Solute carrier family 15 member 2 |
-Macromolecule #2: nanobody
| Macromolecule | Name: nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.494183 KDa |
| Sequence | String: GPSQVQLVES GGGLVQPGGS LRLLCVASGR PFNDYDMGWF RQAPGKEREF VASISWSGRV TDYSDSMKGR CTVSRDNAKG TMFLQMSNL VPRDTAVYYC AAARRRWTFK ATNTEEFYET WGQGTQVTVS SA |
-Macromolecule #3: CLOXACILLIN
| Macromolecule | Name: CLOXACILLIN / type: ligand / ID: 3 / Number of copies: 1 / Formula: CXN |
|---|---|
| Molecular weight | Theoretical: 435.881 Da |
| Chemical component information | 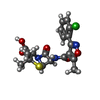 ChemComp-CXN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 57.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)