+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Apo-DIM2 | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA methyltransferase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase activity / negative regulation of gene expression via chromosomal CpG island methylation / methylation / chromatin binding / DNA binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Neurospora crassa (fungus) Neurospora crassa (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Song J / Shao Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation. Authors: Zengyu Shao / Jiuwei Lu / Nelli Khudaverdyan / Jikui Song /  Abstract: Functional crosstalk between DNA methylation, histone H3 lysine-9 trimethylation (H3K9me3) and heterochromatin protein 1 (HP1) is essential for proper heterochromatin assembly and genome stability. ...Functional crosstalk between DNA methylation, histone H3 lysine-9 trimethylation (H3K9me3) and heterochromatin protein 1 (HP1) is essential for proper heterochromatin assembly and genome stability. However, how repressive chromatin cues guide DNA methyltransferases for region-specific DNA methylation remains largely unknown. Here, we report structure-function characterizations of DNA methyltransferase Defective-In-Methylation-2 (DIM2) in Neurospora. The DNA methylation activity of DIM2 requires the presence of both H3K9me3 and HP1. Our structural study reveals a bipartite DIM2-HP1 interaction, leading to a disorder-to-order transition of the DIM2 target-recognition domain that is essential for substrate binding. Furthermore, the structure of DIM2-HP1-H3K9me3-DNA complex reveals a substrate-binding mechanism distinct from that for its mammalian orthologue DNMT1. In addition, the dual recognition of H3K9me3 peptide by the DIM2 RFTS and BAH1 domains allosterically impacts the DIM2-substrate binding, thereby controlling DIM2-mediated DNA methylation. Together, this study uncovers how multiple heterochromatin factors coordinately orchestrate an activity-switching mechanism for region-specific DNA methylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44410.map.gz emd_44410.map.gz | 60 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44410-v30.xml emd-44410-v30.xml emd-44410.xml emd-44410.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44410.png emd_44410.png | 67.4 KB | ||
| Filedesc metadata |  emd-44410.cif.gz emd-44410.cif.gz | 6.7 KB | ||
| Others |  emd_44410_additional_1.map.gz emd_44410_additional_1.map.gz emd_44410_half_map_1.map.gz emd_44410_half_map_1.map.gz emd_44410_half_map_2.map.gz emd_44410_half_map_2.map.gz | 32.4 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44410 http://ftp.pdbj.org/pub/emdb/structures/EMD-44410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44410 | HTTPS FTP |
-Related structure data
| Related structure data |  9bapMC  9baqC  9bazC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44410.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44410.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
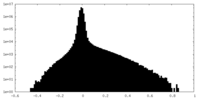
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_44410_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
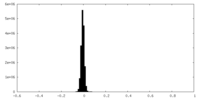
| Density Histograms |
-Half map: half map A
| File | emd_44410_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
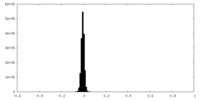
| Density Histograms |
-Half map: half map B
| File | emd_44410_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
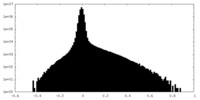
| Density Histograms |
- Sample components
Sample components
-Entire : Apo-DIM2
| Entire | Name: Apo-DIM2 |
|---|---|
| Components |
|
-Supramolecule #1: Apo-DIM2
| Supramolecule | Name: Apo-DIM2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
-Macromolecule #1: DNA (cytosine-5-)-methyltransferase
| Macromolecule | Name: DNA (cytosine-5-)-methyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA (cytosine-5-)-methyltransferase |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 162.352641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMDSPDRSH GGMFIDVPAE TMGFQEDYLD MFASVLSQGL AKEGDYAHHQ PLPAGKEECL EPIAVATTIT PSPDDPQLQL QLELEQQFQ TESGLNGVDP APAPESEDEA DLPDGFSDES PDDDFVVQRS KHITVDLPVS TLINPRSTFQ RIDENDNLVP P PQSTPERV ...String: GSMDSPDRSH GGMFIDVPAE TMGFQEDYLD MFASVLSQGL AKEGDYAHHQ PLPAGKEECL EPIAVATTIT PSPDDPQLQL QLELEQQFQ TESGLNGVDP APAPESEDEA DLPDGFSDES PDDDFVVQRS KHITVDLPVS TLINPRSTFQ RIDENDNLVP P PQSTPERV AVEDLLKAAK AAGKNKEDYI EFELHDFNFY VNYAYHPQEM RPIQLVATKV LHDKYYFDGV LKYGNTKHYV TG MQVLELP VGNYGASLHS VKGQIWVRSK HNAKKEIYYL LKKPAFEYQR YYQPFLWIAD LGKHVVDYCT RMVERKREVT LGC FKSDFI QWASKAHGKS KAFQNWRAQH PSDDFRTSVA ANIGYIWKEI NGVAGAKRAA GDQLFRELMI VKPGQYFRQE VPPG PVVTE GDRTVAATIV TPYIKECFGH MILGKVLRLA GEDAEKEKEV KLAKRLKIEN KNATKADTKD DMKNDTATES LPTPL RSLP VQVLEATPIE SDIVSIVSSD LPPSENNPPP LTNGSVKPKA KANPKPKPST QPLHAAHVKY LSQELVNKIK VGDVIS TPR DDSSNTDTKW KPTDTDDHRW FGLVQRVHTA KTKSSGRGLN SKSFDVIWFY RPEDTPCCAM KYKWRNELFL SNHCTCQ EG HHARVKGNEV LAVHPVDWFG TPESNKGEFF VRQLYESEQR RWITLQKDHL TCYHNQPPKP PTAPYKPGDT VLATLSPS D KFSDPYEVVE YFTQGEKETA FVRLRKLLRR RKVDRQDAPA NELVYTEDLV DVRAERIVGK CIMRCFRPDE RVPSPYDRG GTGNMFFITH RQDHGRCVPL DTLPPTLRQG FNPLGNLGKP KLRGMDLYCG GGNFGRGLEE GGVVEMRWAN DIWDKAIHTY MANTPDPNK TNPFLGSVDD LLRLALEGKF SDNVPRPGEV DFIAAGSPCP GFSLLTQDKK VLNQVKNQSL VASFASFVDF Y RPKYGVLE NVSGIVQTFV NRKQDVLSQL FCALVGMGYQ AQLILGDAWA HGAPQSRERV FLYFAAPGLP LPDPPLPSHS HY RVKNRNI GFLCNGESYV QRSFIPTAFK FVSAGEGTAD LPKIGDGKPD ACVRFPDHRL ASGITPYIRA QYACIPTHPY GMN FIKAWN NGNGVMSKSD RDLFPSEGKT RTSDASVGWK RLNPKTLFPT VTTTSNPSDA RMGPGLHWDE DRPYTVQEMR RAQG YLDEE VLVGRTTDQW KLVGNSVSRH MALAIGLKFR EAWLGTLYDE SAVVATATAT ATTAAAVGVT VPVMEEPGIG TTESS RPSR SPVHTAVDLD DSKSERSRST TPATVLSTSS AAGDGSANAA GLEDDDNDDM EMMEVTRKRS SPAVDEEGMR PSKVQK VEV TVASPASRRS SRQASRNPTA SPSSKASKAT THEAPAPEEL ESDAESYSET YDKEGFDGDY HSGHEDQYSE EDEEEEY AE PETMTVNGMT IVKL UniProtKB: DNA (cytosine-5-)-methyltransferase |
-Macromolecule #2: S-ADENOSYL-L-HOMOCYSTEINE
| Macromolecule | Name: S-ADENOSYL-L-HOMOCYSTEINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: SAH |
|---|---|
| Molecular weight | Theoretical: 384.411 Da |
| Chemical component information |  ChemComp-SAH: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.88 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 529317 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)