[English] 日本語
 Yorodumi
Yorodumi- EMDB-43835: Structure of biofilm-forming functional amyloid PSMa1 from Staphy... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | |||||||||
 Map data Map data | DeepEM processed map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | functional amyloid fibril / biofilm / bacterial biofilm / phenol soluble modulin alpha1 / PSMa1 / STRUCTURAL PROTEIN | |||||||||
| Function / homology | Phenol-soluble modulin alpha peptide / Phenol-soluble modulin alpha peptide family / killing of cells of another organism / Phenol-soluble modulin alpha 1 peptide Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
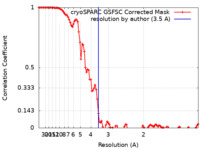
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Hansen KH / Byeon CH / Liu Q / Drace T / Boesen T / Conway JF / Andreasen M / Akbey U | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structure of biofilm-forming functional amyloid PSMα1 from . Authors: Kasper Holst Hansen / Chang Hyeock Byeon / Qian Liu / Taner Drace / Thomas Boesen / James F Conway / Maria Andreasen / Ümit Akbey /   Abstract: Biofilm-protected pathogenic causes chronic infections that are difficult to treat. An essential building block of these biofilms are functional amyloid fibrils that assemble from phenol-soluble ...Biofilm-protected pathogenic causes chronic infections that are difficult to treat. An essential building block of these biofilms are functional amyloid fibrils that assemble from phenol-soluble modulins (PSMs). PSMα1 cross-seeds other PSMs into cross-β amyloid folds and is therefore a key element in initiating biofilm formation. However, the paucity of high-resolution structures hinders efforts to prevent amyloid assembly and biofilm formation. Here, we present a 3.5 Å resolution density map of the major PSMα1 fibril form revealing a left-handed cross-β fibril composed of two C-symmetric U-shaped protofilaments whose subunits are unusually tilted out-of-plane. Monomeric α-helical PSMα1 is extremely cytotoxic to cells, despite the moderate toxicity of the cross-β fibril. We suggest mechanistic insights into the PSM functional amyloid formation and conformation transformation on the path from monomer-to-fibril formation. Details of PSMα1 assembly and fibril polymorphism suggest how utilizes functional amyloids to form biofilms and establish a framework for developing therapeutics against infection and antimicrobial resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43835.map.gz emd_43835.map.gz | 93.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43835-v30.xml emd-43835-v30.xml emd-43835.xml emd-43835.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43835_fsc.xml emd_43835_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_43835.png emd_43835.png | 61 KB | ||
| Masks |  emd_43835_msk_1.map emd_43835_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43835.cif.gz emd-43835.cif.gz | 5.7 KB | ||
| Others |  emd_43835_additional_1.map.gz emd_43835_additional_1.map.gz emd_43835_additional_2.map.gz emd_43835_additional_2.map.gz emd_43835_additional_3.map.gz emd_43835_additional_3.map.gz emd_43835_half_map_1.map.gz emd_43835_half_map_1.map.gz emd_43835_half_map_2.map.gz emd_43835_half_map_2.map.gz | 50.6 MB 52.4 MB 97 MB 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43835 http://ftp.pdbj.org/pub/emdb/structures/EMD-43835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43835 | HTTPS FTP |
-Validation report
| Summary document |  emd_43835_validation.pdf.gz emd_43835_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43835_full_validation.pdf.gz emd_43835_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_43835_validation.xml.gz emd_43835_validation.xml.gz | 18.7 KB | Display | |
| Data in CIF |  emd_43835_validation.cif.gz emd_43835_validation.cif.gz | 23.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43835 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43835 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43835 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43835 | HTTPS FTP |
-Related structure data
| Related structure data |  9atwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43835.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43835.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEM processed map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.647 Å | ||||||||||||||||||||||||||||||||||||
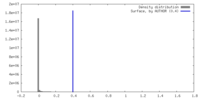
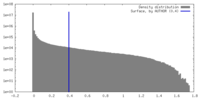
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43835_msk_1.map emd_43835_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
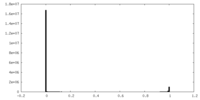
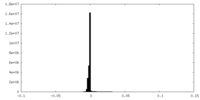
| Density Histograms |
-Additional map: CryoSparc output unsharpened.
| File | emd_43835_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc output unsharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
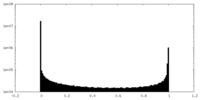
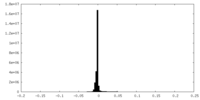
| Density Histograms |
-Additional map: CryoSparc sharpened to B:-100
| File | emd_43835_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc sharpened to B:-100 | ||||||||||||
| Projections & Slices |
| ||||||||||||
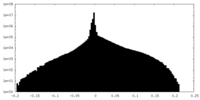
| Density Histograms |
-Additional map: CryoSparc auto-sharpened
| File | emd_43835_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc auto-sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
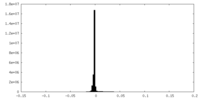
| Density Histograms |
-Half map: Half Map B from CryoSparc
| File | emd_43835_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B from CryoSparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A from CryoSparc
| File | emd_43835_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A from CryoSparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phenol Soluble Modulin alpha1 (PSMAlpha1) (PSMa1)
| Entire | Name: Phenol Soluble Modulin alpha1 (PSMAlpha1) (PSMa1) |
|---|---|
| Components |
|
-Supramolecule #1: Phenol Soluble Modulin alpha1 (PSMAlpha1) (PSMa1)
| Supramolecule | Name: Phenol Soluble Modulin alpha1 (PSMAlpha1) (PSMa1) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Biofilm forming functional amyloid from Staphylococcus aureus PSMa1 is produced by peptide-synthesis |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Phenol-soluble modulin alpha 1 peptide
| Macromolecule | Name: Phenol-soluble modulin alpha 1 peptide / type: protein_or_peptide / ID: 1 / Number of copies: 64 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.262817 KDa |
| Sequence | String: MGIIAGIIKV IKSLIEQFTG K UniProtKB: Phenol-soluble modulin alpha 1 peptide |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: water |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. Details: C-Flat R2/2 Cu 300 mesh holey carbon grids (Protochips) were glow discharged for 45 s at 15 mA using a Quorum GloQube Plus |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 99 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: plunge-frozen in liquid ethane using a Vitrobot Mark IV plunge freezer with a blot force of zero at 4C and 99% humidity.. |
| Details | 3 microL of PSMa1 (0.5 mg/mL) was applied to grids Blotted for 6s |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3002 / Average exposure time: 1.5 sec. / Average electron dose: 63.0 e/Å2 Details: A total of 3,002 movies of the PSMa1 sample were collected |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT / Overall B value: 50 |
|---|---|
| Output model |  PDB-9atw: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)