[English] 日本語
 Yorodumi
Yorodumi- EMDB-43751: TRPM7 structure in complex with anticancer agent CCT128930 in clo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | transient receptor potential M family member 7 / TRP / channel / TRPM7 / TRP channels / membrane protein / CCT128930 | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationintracellular magnesium ion homeostasis / calcium-dependent cell-matrix adhesion / monoatomic cation homeostasis / varicosity / TRP channels / actomyosin structure organization / myosin binding / monoatomic cation transmembrane transport / necroptotic process / monoatomic cation channel activity ...intracellular magnesium ion homeostasis / calcium-dependent cell-matrix adhesion / monoatomic cation homeostasis / varicosity / TRP channels / actomyosin structure organization / myosin binding / monoatomic cation transmembrane transport / necroptotic process / monoatomic cation channel activity / ruffle / bioluminescence / generation of precursor metabolites and energy / protein tetramerization / calcium channel activity / memory / synaptic vesicle membrane / calcium ion transport / kinase activity / actin binding / non-specific serine/threonine protein kinase / protein kinase activity / positive regulation of apoptotic process / protein serine kinase activity / protein serine/threonine kinase activity / neuronal cell body / ATP binding / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.45 Å | ||||||||||||||||||
 Authors Authors | Nadezhdin KD / Sobolevsky AI | ||||||||||||||||||
| Funding support |  United States, United States,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930. Authors: Kirill D Nadezhdin / Leonor Correia / Alexey Shalygin / Muhammed Aktolun / Arthur Neuberger / Thomas Gudermann / Maria G Kurnikova / Vladimir Chubanov / Alexander I Sobolevsky /   Abstract: TRP channels are implicated in various diseases, but high structural similarity between them makes selective pharmacological modulation challenging. Here, we study the molecular mechanism underlying ...TRP channels are implicated in various diseases, but high structural similarity between them makes selective pharmacological modulation challenging. Here, we study the molecular mechanism underlying specific inhibition of the TRPM7 channel, which is essential for cancer cell proliferation, by the anticancer agent CCT128930 (CCT). Using cryo-EM, functional analysis, and MD simulations, we show that CCT binds to a vanilloid-like (VL) site, stabilizing TRPM7 in the closed non-conducting state. Similar to other allosteric inhibitors of TRPM7, NS8593 and VER155008, binding of CCT is accompanied by displacement of a lipid that resides in the VL site in the apo condition. Moreover, we demonstrate the principal role of several residues in the VL site enabling CCT to inhibit TRPM7 without impacting the homologous TRPM6 channel. Hence, our results uncover the central role of the VL site for the selective interaction of TRPM7 with small molecules that can be explored in future drug design. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43751.map.gz emd_43751.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43751-v30.xml emd-43751-v30.xml emd-43751.xml emd-43751.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43751_fsc.xml emd_43751_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_43751.png emd_43751.png | 242.1 KB | ||
| Filedesc metadata |  emd-43751.cif.gz emd-43751.cif.gz | 7.1 KB | ||
| Others |  emd_43751_half_map_1.map.gz emd_43751_half_map_1.map.gz emd_43751_half_map_2.map.gz emd_43751_half_map_2.map.gz | 115.2 MB 115.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43751 http://ftp.pdbj.org/pub/emdb/structures/EMD-43751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43751 | HTTPS FTP |
-Validation report
| Summary document |  emd_43751_validation.pdf.gz emd_43751_validation.pdf.gz | 804 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43751_full_validation.pdf.gz emd_43751_full_validation.pdf.gz | 803.6 KB | Display | |
| Data in XML |  emd_43751_validation.xml.gz emd_43751_validation.xml.gz | 17.3 KB | Display | |
| Data in CIF |  emd_43751_validation.cif.gz emd_43751_validation.cif.gz | 22.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43751 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43751 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43751 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43751 | HTTPS FTP |
-Related structure data
| Related structure data |  8w2lMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43751.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43751.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43751_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
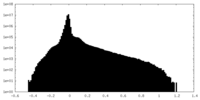
| Projections & Slices |
| ||||||||||||
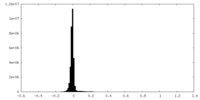
| Density Histograms |
-Half map: #1
| File | emd_43751_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
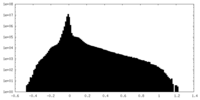
| Projections & Slices |
| ||||||||||||
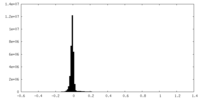
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of TRPM7 with antagonist CCT128930
| Entire | Name: Complex of TRPM7 with antagonist CCT128930 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of TRPM7 with antagonist CCT128930
| Supramolecule | Name: Complex of TRPM7 with antagonist CCT128930 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Green fluorescent protein,Transient receptor potential cation cha...
| Macromolecule | Name: Green fluorescent protein,Transient receptor potential cation channel subfamily M member 7 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 175.75625 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MWSHPQFEKV SKGEELFTGV VPILVELDGD VNGHKFSVSG EGEGDATYGK LTLKFICTTG KLPVPWPTLV TTLTYGVQCF SRYPDHMKQ HDFFKSAMPE GYVQERTIFF KDDGNYKTRA EVKFEGDTLV NRIELKGIDF KEDGNILGHK LEYNYNSHNV Y IMADKQKN ...String: MWSHPQFEKV SKGEELFTGV VPILVELDGD VNGHKFSVSG EGEGDATYGK LTLKFICTTG KLPVPWPTLV TTLTYGVQCF SRYPDHMKQ HDFFKSAMPE GYVQERTIFF KDDGNYKTRA EVKFEGDTLV NRIELKGIDF KEDGNILGHK LEYNYNSHNV Y IMADKQKN GIKVNFKIRH NIEDGSVQLA DHYQQNTPIG DGPVLLPDNH YLSTQSKLSK DPNEKRDHMV LLEFVTAAGI TL GMDELYK VDLVPRGSAQ KSWIESTLTK RECVYIIPSS KDPHRCLPGC QICQQLVRCF CGRLVKQHAC FTASLAMKYS DVK LGEHFN QAIEEWSVEK HTEQSPTDAY GVINFQGGSH SYRAKYVRLS YDTKPEIILQ LLLKEWQMEL PKLVISVHGG MQKF ELHPR IKQLLGKGLI KAAVTTGAWI LTGGVNTGVA KHVGDALKEH ASRSSRKICT IGIAPWGVIE NRNDLVGRDV VAPYQ TLLN PLSKLNVLNN LHSHFILVDD GTVGKYGAEV RLRRELEKTI NQQRIHARIG QGVPVVALIF EGGPNVILTV LEYLQE SPP VPVVVCEGTG RAADLLAYIH KQTEEGGNLP DAAEPDIIST IKKTFNFGQS EAVHLFQTMM ECMKKKELIT VFHIGSE DH QDIDVAILTA LLKGTNASAF DQLILTLAWD RVDIAKNHVF VYGQQWLVGS LEQAMLDALV MDRVSFVKLL IENGVSMH K FLTIPRLEEL YNTKQGPTNP MLFHLIRDVK QGNLPPGYKI TLIDIGLVIE YLMGGTYRCT YTRKRFRLIY NSLGGNNRR SGRNTSSSTP QLRKSHETFG NRADKKEKMR HNHFIKTAQP YRPKMDASME EGKKKRTKDE IVDIDDPETK RFPYPLNELL IWACLMKRQ VMARFLWQHG EESMAKALVA CKIYRSMAYE AKQSDLVDDT SEELKQYSND FGQLAVELLE QSFRQDETMA M KLLTYELK NWSNSTCLKL AVSSRLRPFV AHTCTQMLLS DMWMGRLNMR KNSWYKVILS ILVPPAILML EYKTKAEMSH IP QSQDAHQ MTMEDSENNF HNITEEIPME VFKEVKILDS SDGKNEMEIH IKSKKLPITR KFYAFYHAPI VKFWFNTLAY LGF LMLYTF VVLVKMEQLP SVQEWIVIAY IFTYAIEKVR EVFMSEAGKI SQKIKVWFSD YFNVSDTIAI ISFFVGFGLR FGAK WNYIN AYDNHVFVAG RLIYCLNIIF WYVRLLDFLA VNQQAGPYVM MIGKMVANMF YIVVIMALVL LSFGVPRKAI LYPHE EPSW SLAKDIVFHP YWMIFGEVYA YEIDVCANDS TLPTICGPGT WLTPFLQAVY LFVQYIIMVN LLIAFFNNVY LQVKAI SNI VWKYQRYHFI MAYHEKPVLP PPLIILSHIV SLFCCVCKRR KKDKTSDGPK LFLTEEDQKK LHDFEEQCVE MYFDEKD DK FNSGSEERIR VTFERVEQMS IQIKEVGDRV NYIKRSLQSL DSQIGHLQDL SALTVDTLKT LTAQKASEAS KVHNEITR E LSISKHLAQN LID UniProtKB: Green fluorescent protein, Transient receptor potential cation channel subfamily M member 7 |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 52 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 4 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4...
| Macromolecule | Name: 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium type: ligand / ID: 4 / Number of copies: 4 / Formula: M05 |
|---|---|
| Molecular weight | Theoretical: 342.846 Da |
| Chemical component information | 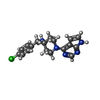 ChemComp-M05: |
-Macromolecule #5: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13...
| Macromolecule | Name: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol type: ligand / ID: 5 / Number of copies: 4 / Formula: DU0 |
|---|---|
| Molecular weight | Theoretical: 516.752 Da |
| Chemical component information |  ChemComp-DU0: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 164 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.9 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)