[English] 日本語
 Yorodumi
Yorodumi- EMDB-43737: Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Streptomyces / Umbrella toxin particles / Alanine leucine phenylalanine-rich (ALF) repeat proteins / Seattle structural genomics center for infectious disease / SSGCID / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Park YJ / Zhao Q / Seattle Structural Genomics Center for Infectious Disease (SSGCID) / DiMaio F / Mougous JD / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Streptomyces umbrella toxin particles block hyphal growth of competing species. Authors: Qinqin Zhao / Savannah Bertolli / Young-Jun Park / Yongjun Tan / Kevin J Cutler / Pooja Srinivas / Kyle L Asfahl / Citlali Fonesca-García / Larry A Gallagher / Yaqiao Li / Yaxi Wang / Devin ...Authors: Qinqin Zhao / Savannah Bertolli / Young-Jun Park / Yongjun Tan / Kevin J Cutler / Pooja Srinivas / Kyle L Asfahl / Citlali Fonesca-García / Larry A Gallagher / Yaqiao Li / Yaxi Wang / Devin Coleman-Derr / Frank DiMaio / Dapeng Zhang / S Brook Peterson / David Veesler / Joseph D Mougous /  Abstract: Streptomyces are a genus of ubiquitous soil bacteria from which the majority of clinically utilized antibiotics derive. The production of these antibacterial molecules reflects the relentless ...Streptomyces are a genus of ubiquitous soil bacteria from which the majority of clinically utilized antibiotics derive. The production of these antibacterial molecules reflects the relentless competition Streptomyces engage in with other bacteria, including other Streptomyces species. Here we show that in addition to small-molecule antibiotics, Streptomyces produce and secrete antibacterial protein complexes that feature a large, degenerate repeat-containing polymorphic toxin protein. A cryo-electron microscopy structure of these particles reveals an extended stalk topped by a ringed crown comprising the toxin repeats scaffolding five lectin-tipped spokes, which led us to name them umbrella particles. Streptomyces coelicolor encodes three umbrella particles with distinct toxin and lectin composition. Notably, supernatant containing these toxins specifically and potently inhibits the growth of select Streptomyces species from among a diverse collection of bacteria screened. For one target, Streptomyces griseus, inhibition relies on a single toxin and that intoxication manifests as rapid cessation of vegetative hyphal growth. Our data show that Streptomyces umbrella particles mediate competition among vegetative mycelia of related species, a function distinct from small-molecule antibiotics, which are produced at the onset of reproductive growth and act broadly. Sequence analyses suggest that this role of umbrella particles extends beyond Streptomyces, as we identified umbrella loci in nearly 1,000 species across Actinobacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43737.map.gz emd_43737.map.gz | 101.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43737-v30.xml emd-43737-v30.xml emd-43737.xml emd-43737.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43737.png emd_43737.png | 48.8 KB | ||
| Filedesc metadata |  emd-43737.cif.gz emd-43737.cif.gz | 6.6 KB | ||
| Others |  emd_43737_additional_1.map.gz emd_43737_additional_1.map.gz emd_43737_half_map_1.map.gz emd_43737_half_map_1.map.gz emd_43737_half_map_2.map.gz emd_43737_half_map_2.map.gz | 54.2 MB 99.6 MB 99.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43737 http://ftp.pdbj.org/pub/emdb/structures/EMD-43737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43737 | HTTPS FTP |
-Related structure data
| Related structure data |  8w22MC  8w20C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43737.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43737.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.686 Å | ||||||||||||||||||||||||||||||||||||
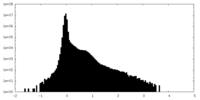
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_43737_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
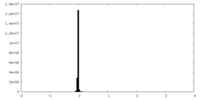
| Density Histograms |
-Half map: #2
| File | emd_43737_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
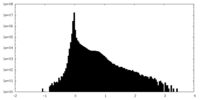
| Density Histograms |
-Half map: #1
| File | emd_43737_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
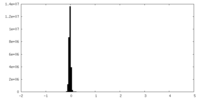
| Density Histograms |
- Sample components
Sample components
-Entire : Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF...
| Entire | Name: Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) |
|---|---|
| Components |
|
-Supramolecule #1: Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF...
| Supramolecule | Name: Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
-Macromolecule #1: Intein C-terminal splicing domain-containing protein
| Macromolecule | Name: Intein C-terminal splicing domain-containing protein / type: protein_or_peptide / ID: 1 Details: endogenous tags to purified the complex from natural source Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Molecular weight | Theoretical: 142.716734 KDa |
| Sequence | String: MRRRIPSRTP GSGAKQKSWF PRRSLQVLLS AGMLSGLLGT PAALAAEPAP LPANVRADIV GYWETGGAGL KAAAEQALLG GDEAIRKFL ADAPSIQHDD NRIDAARMAM TGGSGLRQAA KDAIRLSPAE LEKFLLYGYE EPLDDDHKVE IARLINLGGP G VREAGKAA ...String: MRRRIPSRTP GSGAKQKSWF PRRSLQVLLS AGMLSGLLGT PAALAAEPAP LPANVRADIV GYWETGGAGL KAAAEQALLG GDEAIRKFL ADAPSIQHDD NRIDAARMAM TGGSGLRQAA KDAIRLSPAE LEKFLLYGYE EPLDDDHKVE IARLINLGGP G VREAGKAA LQGTAEEREL FLNSGQYTAQ QDDNRVDVAR LATTGGPNVQ AAAKVALRGT PEDMVEFLEV GQFTARNRDQ EH ATIAELI KQAELAGKQA DDARKTAEES SKKAVDASQL AKEAAQKAAE ETEAAKDDSQ KAAVKAKQAA DAARAAADAA QEA IGSANA ANRAARRAAL AAAQTASAAT AAAEAANKAY KAAIAAAGDE GKADEAKEAA KQARAAADAA TKSGLAAENA GLAS AAAAT ASTAAKSASS NARAAAGAAE EANQHADAAG VHSNEAALAA AEARRHADAA DRAADRSSAL AQRAATAAYG ARDAA NSAA EHANKAADYA DESAAHAGDS AAYAATAKRN AQAAQEAAKT ATAAVTKANE IFKLARETET ANLETRTDAA IERARS MKS ASETSITASA TTQVEALALN DTATELAKEA SRPDIDVQAT AAKGRQLAMQ AMKLLGPWHQ EAAARALSGT DQDVLDY LR TRWKEANHND IRQQIVDLST QSPYASVRTA AAEALNGTPE QIEAFHTTGQ YTAGSDDMKV DVARLANTGG PGVSQAAK T ALADGTGKTL ATFLQIGQYG ERLSDEKVVT ARLAETGSPE VQAAAKIALA GPPELLHEFV TTGQYMAKRK DDLADVHVN QVERLLAEGS LIAAEANEDA WRATEAAATA EGAAADAATA AEKAEASAAQ AKQHAADADA SADAATRSAA DAAASAATAR DAADRAAQD ATAAENSAAE AAFSAAYARD SASKADDAAD RARASALAAG KSADEAEAEA KEAWKTTRAL AEKEAEEARR K AAEERKRQ QEQAGEPKRV CIPHPTRETM IPIMPCAASP DDSMIMPGPV DPTIRAVVWE LAGLNDIKAC IDKPLSGNCV MA VVGVTPW GKFKLVSKLG NGLDAVKDAR GARRTVACLT GAAHSFPAGT RVLMADGTRR SIEQIEAGDL VTATDPTTGE TGA RTVTRT IHTPDDRNFT DVALADGSTL TSTTHHPYWS QNDQTWKNAG DLEAGDTLRT PQNTAVVIAA THDWPGLQDA YDLT VDGFH SYYVSTGTTD VLVHNNDNPC PDWVSKAWKK LPKRKSGDPT SGYVFEADGT LVWDSVLTSG RSPLSEDISA FLKGS PDFP NFPGYADVAH HAEAKIAWEM RTKMGKGKKL HIVINTNYVC PKVSSPNSMG CKQAVPAILY EDQTLYVHYP GASDAL ELK GTAKR UniProtKB: Hint domain-containing protein |
-Macromolecule #2: Secreted protein
| Macromolecule | Name: Secreted protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Molecular weight | Theoretical: 17.18627 KDa |
| Sequence | String: MANTSRTRQA LMAIAVSVLA AGVTTLGVAH ADNGDAVAAA AEMPQAVEDF SYPGAAKIQA ETGAILKRGN GHMLMTSCDG SEDIQVMSR TGQKDFCFNV MAKPAYLTLE VPQAYGIWTS ADPVKTTIKD TDGTATVINA PANDFTGYGE AGSTGEPTTL I ELRVAG UniProtKB: Secreted protein |
-Macromolecule #3: Secreted esterase
| Macromolecule | Name: Secreted esterase / type: protein_or_peptide / ID: 3 Details: endogenous tags to purified the complex from natural source Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Molecular weight | Theoretical: 54.437102 KDa |
| Sequence | String: MFRMPRPIRA TALSAAVLAG ALASTPAQAT VGDPTTDAKL DFTARLTIGT DYRSCSGALV DTQWVLTAAS CFADDPNQPD TVAAGKPAQ LTRATVGRAD SNIANGYVRE VVELVPHPER DMVLARLDKA IPDIAPVRLA SDAPTAGTPL TAVGFGRTKD E WVPIQRHQ ...String: MFRMPRPIRA TALSAAVLAG ALASTPAQAT VGDPTTDAKL DFTARLTIGT DYRSCSGALV DTQWVLTAAS CFADDPNQPD TVAAGKPAQ LTRATVGRAD SNIANGYVRE VVELVPHPER DMVLARLDKA IPDIAPVRLA SDAPTAGTPL TAVGFGRTKD E WVPIQRHQ GAFTVTSVTA GAVNVTGQGG DAICAGDTGG PLLQDKNGTL HLVGVNNRSM QGGCYGSETT STDAIAAMSD AD FVTQTVN RDLGTGNLSD LVASADFNSD GRTDVAAVLE DGSLHAFYAK PDGTLEYGRE LWNDNTWSPM VQIIGGDFNS DGN GDIAAV RSDGTLNLYT GTATGILNKS KPMWHDTSWK TIKQVTRFKF NGRDGLVAQW GDGNLYGYYT GTDGTLTGTK VKMW PDATW GKTRLTGTAD INADGRDDLT AVRDDGSLNW YAGNTKGGLD AARKLWPDNT WTPMKRIIGG DFNGDNKGDI AAVGG QSTL LLYTGTGTGT LNKGIAMRPA SGSHHHHHHH H UniProtKB: Secreted esterase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 386275 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)